7BXZ
 
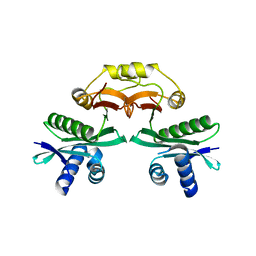 | |
7CP1
 
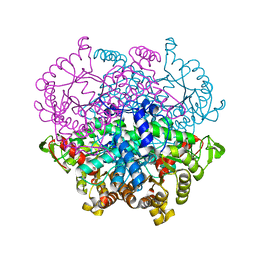 | |
2N8E
 
 | |
5IJA
 
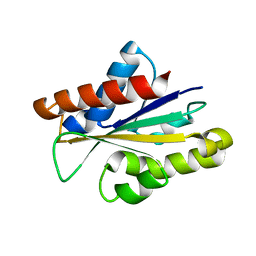 | |
5YY0
 
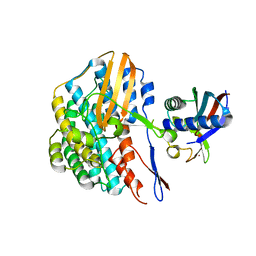 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6L8P
 
 | | Crystal structure of RidA from Antarctic bacterium Psychrobacter sp. PAMC 21119 | | Descriptor: | MALONATE ION, RidA family protein | | Authors: | Kwon, S, Lee, C.W, Koh, H.Y, Lee, J.H, Park, H.H. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of the reactive intermediate/imine deaminase A homolog from the Antarctic bacterium Psychrobacter sp. PAMC 21119.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
5ZBY
 
 | |
2Y5Y
 
 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6IO1
 
 | |
6J52
 
 | | Crystal structure of CARD-only protein in frog virus 3 | | Descriptor: | Caspase recruitment domain-only protein | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural transformation-mediated dimerization of caspase recruitment domain revealed by the crystal structure of CARD-only protein in frog virus 3.
J. Struct. Biol., 205, 2019
|
|
6IZ9
 
 | |
7VPF
 
 | |
6A8P
 
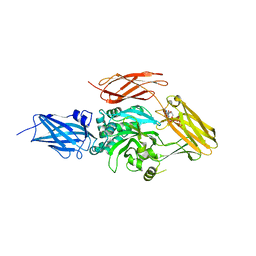 | | Transglutaminase 2 mutant G224V in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Park, H.H, Ha, H.J, Kwon, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity.
PLoS ONE, 13, 2018
|
|
8TK7
 
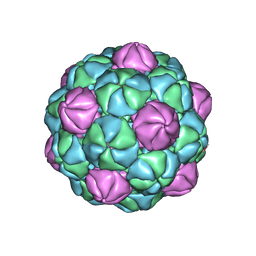 | |
6K8H
 
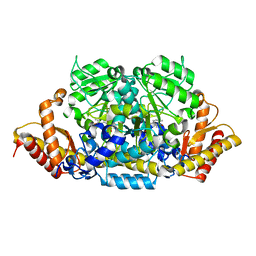 | | Crystal structure of an omega-transaminase from Sphaerobacter thermophilus | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Aminotransferase class-III | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the enzyme specificity of a novel omega-transaminase from the thermophilic bacterium Sphaerobacter thermophilus.
J.Struct.Biol., 208, 2019
|
|
5WOV
 
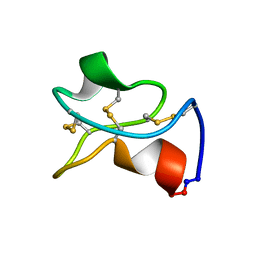 | |
5WOW
 
 | |
