1GQM
 
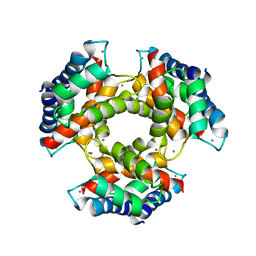 | | The structure of S100A12 in a hexameric form and its proposed role in receptor signalling | | Descriptor: | CALCIUM ION, CALGRANULIN C | | Authors: | Moroz, O.V, Antson, A.A, Dodson, E.G, Burrel, H.J, Grist, S.J, Lloyd, R.M, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E, Bronstein, I.B. | | Deposit date: | 2001-11-26 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of S100A12 in a Hexameric Form and its Proposed Role in Receptor Signalling
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7UR2
 
 | |
8PC7
 
 | |
1VYQ
 
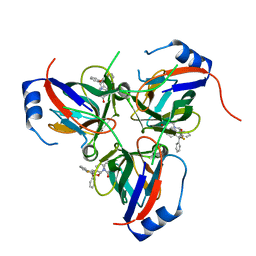 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
7PX3
 
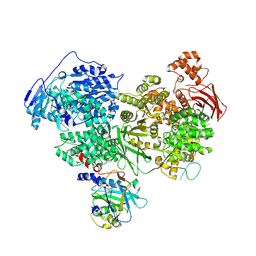 | | Structure of U5 snRNP assembly and recycling factor TSSC4 in complex with BRR2 and Jab1 domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein TSSC4, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Kuropka, B, Ilik, I.A, Freund, C, Aktas, T, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
5EHL
 
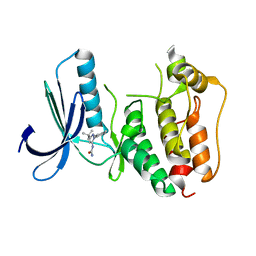 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Dual specificity protein kinase TTK | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
5M13
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, synthetic Nanobody L2_C06 (a-MBP#2) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.372 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
5NJ3
 
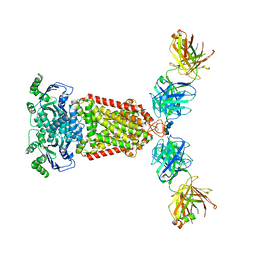 | | Structure of an ABC transporter: complete structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
7RKL
 
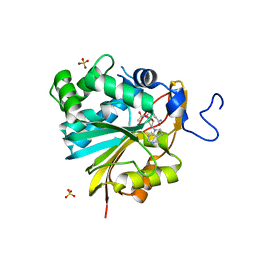 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (P1 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein, SULFATE ION | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2LEJ
 
 | | human prion protein mutant HuPrP(90-231, M129, V210I) | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Raspadori, A, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Toward the Molecular Basis of Inherited Prion Diseases: NMR Structure of the Human Prion Protein with V210I Mutation.
J.Mol.Biol., 412, 2011
|
|
4LHM
 
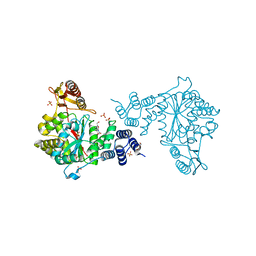 | | Thymidine phosphorylase from E.coli with 3'-azido-3'-deoxythymidine | | Descriptor: | 3'-azido-3'-deoxythymidine, GLYCEROL, SULFATE ION, ... | | Authors: | Timofeev, V.I, Abramchik, Y.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2013-07-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | 3'-Azidothymidine in the active site of Escherichia coli thymidine phosphorylase: the peculiarity of the binding on the basis of X-ray study.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1HB2
 
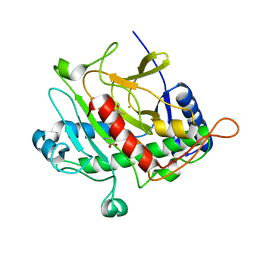 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN EXPOSED PRODUCT FROM ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1S)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOCARBONYL)-2-OXOETHYL]-6-OXO-L-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
7UVA
 
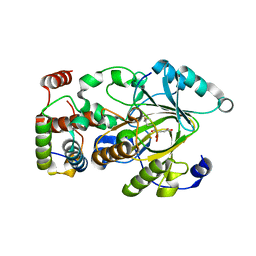 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
6B65
 
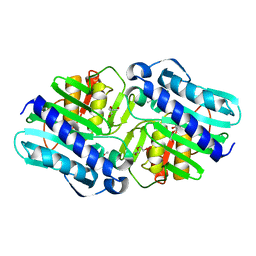 | | IMPase (AF2372) R92Q/K164E | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, ISOPROPYL ALCOHOL, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
6GN3
 
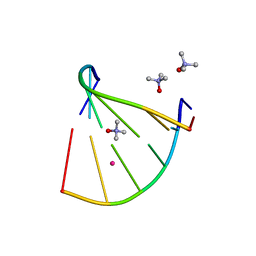 | | Racemic crystal structure of A-DNA duplex formed from d(CCCGGG) in space group P21/n | | Descriptor: | CHLORIDE ION, COBALT (II) ION, DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), ... | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2018-05-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Racemic crystal structures of A-DNA duplexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6B6G
 
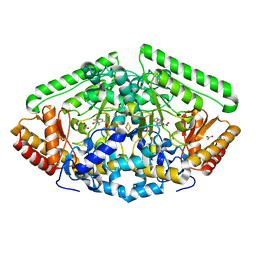 | | Crystal Structure of GABA Aminotransferase bound to (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic acid, an Potent Inactivatorfor the Treatment of Addiction | | Descriptor: | (3R,4E)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]cyclopent-1-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Mascarenhas, R, Juncosa, J.I, Takaya, K, Le, L.V, Moschitto, M.J, Silverman, R.B, Liu, D. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Mechanism of (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic Acid, a Highly Potent gamma-Aminobutyric Acid Aminotransferase Inactivator for the Treatment of Addiction.
J. Am. Chem. Soc., 140, 2018
|
|
5NKU
 
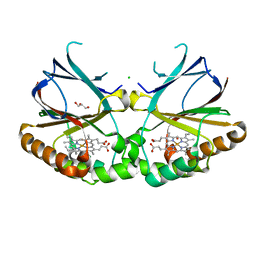 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
8D06
 
 | | Hallucinated C3 protein assembly HALC3_104 | | Descriptor: | HALC3_104 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
5N69
 
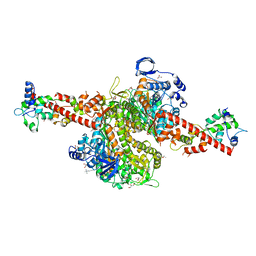 | | Cardiac muscle myosin S1 fragment in the pre-powerstroke state co-crystallized with the activator Omecamtiv Mecarbil | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Planelles-Herrero, V.J, Hartman, J.J, Robert-Paganin, J, Malik, F.I, Houdusse, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil.
Nat Commun, 8, 2017
|
|
1VZ4
 
 | | Fe-Succinate Complex of AtsK | | Descriptor: | FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
5N2V
 
 | |
7PTB
 
 | |
5WAU
 
 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Synchrotron X-Ray Crystallography at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Fromme, R, Ishigami, I, Yeh, S.Y, Zatsepin, N, Grant, T, Fromme, P, Rousseau, D. | | Deposit date: | 2017-06-27 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7PT5
 
 | |
6Y49
 
 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
