3I6O
 
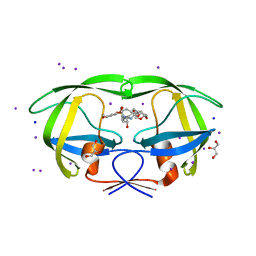 | | Crystal structure of wild type HIV-1 protease with macrocyclic inhibitor GRL-0216A | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-2-hydroxy-3-[(7E)-13-methoxy-1,1-dioxido-3,4,5,6,9,10-hexahydro-2H-11,1,2-benzoxathiazacyclotridecin-2-yl]propyl}carbamate, GLYCEROL, IODIDE ION, ... | | Authors: | Chumanevich, A.A, Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
1RBM
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
6LX4
 
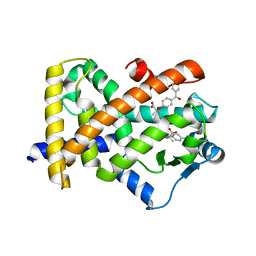 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
1YIK
 
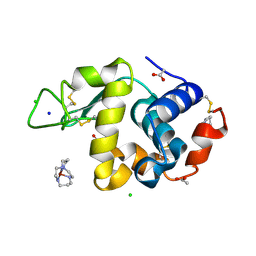 | | Structure of Hen egg white lysozyme soaked with Cu-cyclam | | Descriptor: | 1,4,8,11-TETRAAZA-CYCLOTETRADECANE CU(II), ACETATE ION, CHLORIDE ION, ... | | Authors: | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6AW8
 
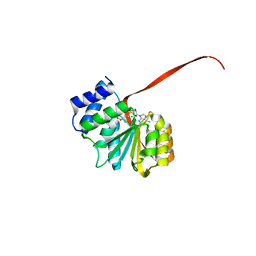 | | 2.25A resolution domain swapped dimer structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
4UHB
 
 | | Laboratory evolved variant R-C1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,2-ETHANEDIOL, EPOXIDE HYDROLASE, GLYCEROL | | Authors: | Nilsson, M.T.I, Carlsson, A.J, Dobritzsch, D, Widersten, M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
6AVN
 
 | |
1RBY
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
6B6I
 
 | | 2.4A resolution structure of human Norovirus GII.4 protease | | Descriptor: | 3C-like protease | | Authors: | Muzzarelli, K.M, Kuiper, B.D, Spellmon, N.S, Hackett, J, Brunzelle, J.S, Kovari, I.A, Amblard, F, Yang, Z, Schinazi, R.F, Kovari, L.C. | | Deposit date: | 2017-10-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and Antiviral Studies of the Human Norovirus GII.4 Protease.
Biochemistry, 58, 2019
|
|
6AOP
 
 | |
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6B84
 
 | |
4UPF
 
 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
6AOS
 
 | |
4O5N
 
 | | Crystal structure of A/Victoria/361/2011 (H3N2) influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
7MIO
 
 | | Mouse TRPV3 in cNW11 nanodiscs, open state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NIY
 
 | | E. coli NfsA with FMN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NB9
 
 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
7NNX
 
 | | E. coli NfsA with 1,4-benzoquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, CHLORIDE ION, ... | | Authors: | Day, M.D, Jarrom, D, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
9B37
 
 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
6AOQ
 
 | |
7NMP
 
 | | E. coli NfsA with hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.D, Jarrom, D, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
1YJ7
 
 | | Crystal structure of enteropathogenic E.coli (EPEC) type III secretion system protein EscJ | | Descriptor: | GLYCEROL, PHOSPHATE ION, escJ | | Authors: | Yip, C.K, Kimbrough, T.G, Felise, H.B, Vuckovic, M, Thomas, N.A, Pfuetzner, R.A, Frey, E.A, Finlay, B.B, Miller, S.I, Strynadka, N.C.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the molecular platform for type III secretion system assembly.
Nature, 435, 2005
|
|
4U14
 
 | | Structure of the M3 muscarinic acetylcholine receptor bound to the antagonist tiotropium crystallized with disulfide-stabilized T4 lysozyme (dsT4L) | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3,Endolysin,Muscarinic acetylcholine receptor M3 | | Authors: | Thorsen, T.S, Matt, R.A, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2014-07-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
6AOU
 
 | |
