2BVX
 
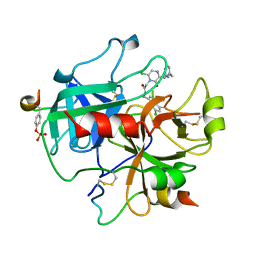 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | ALPHA THROMBIN, HIRUDIN VARIANT-2, N-(5-CHLORO-BENZO[B]THIOPHEN-3-YLMETHYL)-2-[6-CHLORO-OXO-3-(2-PYRIDIN-2-YL-ETHYLAMINO)-2H-PYRAZIN-1-YL]-ACETAMIDE | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
1PXP
 
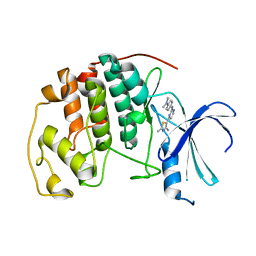 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-N',N'-dimethyl-benzene-1,4-diamine | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-N',N'-DIMETHYL-BENZENE-1,4-DIAMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
5Y9T
 
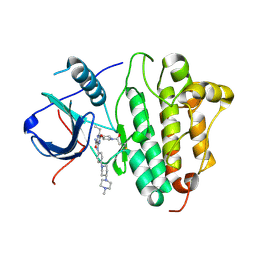 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | Descriptor: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | Deposit date: | 2017-08-28 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
8KDL
 
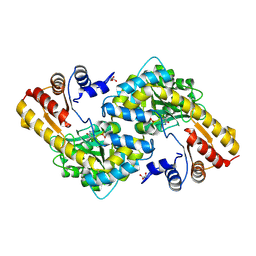 | |
1GU1
 
 | | Crystal structure of type II dehydroquinase from Streptomyces coelicolor complexed with 2,3-anhydro-quinic acid | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Roszak, A.W, Robinson, D.A, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
7SKQ
 
 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor GRL-0617 | | Descriptor: | 3C-like proteinase, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ZINC ION | | Authors: | Freitas, B, Durie, I, Shepard, J, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
8KIC
 
 | | Bacterial serine protease | | Descriptor: | Lysozyme fragment (unknown sequence), peptidase Do | | Authors: | Lee, I.-G, Jeon, H. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of bacterial serine protease
To Be Published
|
|
5NII
 
 | | Crystal structure of the atypical thioredoxin reductase TRi from Desulfovibrio vulgaris Hildenborough | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Valette, O, Tran, T.T.I, Cavazza, C, Caudeville, E, Brasseur, G, Dolla, A, Talla, E, Pieulle, L. | | Deposit date: | 2017-03-24 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Function, Molecular Structure and Evolution of an Atypical Thioredoxin Reductase from Desulfovibrio vulgaris.
Front Microbiol, 8, 2017
|
|
7QF1
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with the human antibody CV2.6264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV2.6264 heavy chain, CV2.6264 light chain, ... | | Authors: | Fernandez, I, Pederzoli, R, Rey, F.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2.
J.Exp.Med., 219, 2022
|
|
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
7QEZ
 
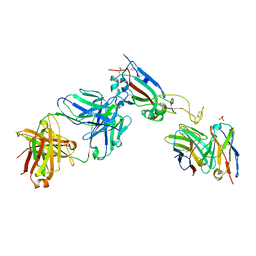 | |
7QVE
 
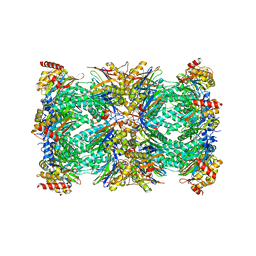 | | Spinach 20S proteasome | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit alpha type-3, Proteasome subunit beta, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome.
Plant Commun., 3, 2022
|
|
8OO0
 
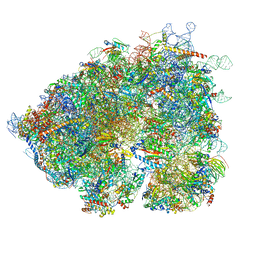 | | Chaetomium thermophilum Methionine Aminopeptidase 2 autoproteolysis product at the 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0, ... | | Authors: | Klein, M.A, Wild, K, Kisonaite, M, Sinning, I. | | Deposit date: | 2023-04-04 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Methionine aminopeptidase 2 and its autoproteolysis product have different binding sites on the ribosome.
Nat Commun, 15, 2024
|
|
5YP4
 
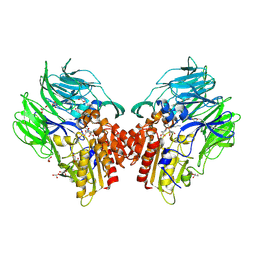 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
6XMP
 
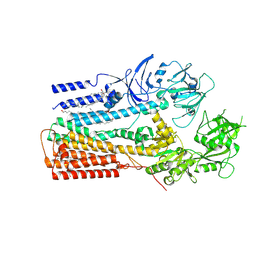 | | Structure of P5A-ATPase Spf1, Apo form | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6XPP
 
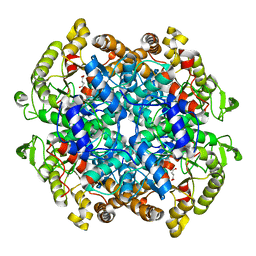 | |
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
2Q9G
 
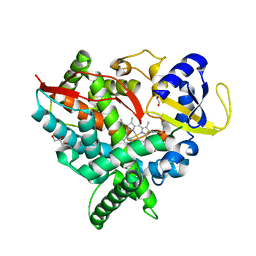 | | Crystal structure of human cytochrome P450 46A1 | | Descriptor: | Cytochrome P450 46A1, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5NOR
 
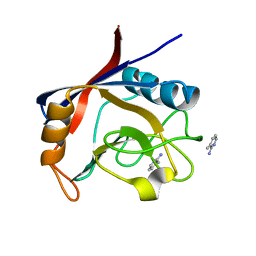 | | Structure of cyclophilin A in complex with 3-methylpyridin-2-amine | | Descriptor: | 3-methylpyridin-2-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
6YOD
 
 | | Structure of Lysozyme from SiN IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(2-METHOXYETHOXY)ETHANOL, ACETIC ACID, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
5NOY
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzamide | | Descriptor: | 3,4-bis(azanyl)benzamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
8P0S
 
 | | Crystal structure HR1 domain of Rho-associated coiled-coil protein kinases (ROCK-HR1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Rho-associated protein kinase 1 | | Authors: | Dubey, B.N, Dvorsky, R, Gremer, L, Vetter, I.R, Schmitt, L, Groth, G, Ahmadian, M.R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the p160 Rho-associated coiled-coil-containing protein kinase
To Be Published
|
|
2Q7Y
 
 | | Structure of the endogenous iNKT cell ligand iGb3 bound to mCD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Wilson, I.A, Teyton, L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
1IFB
 
 | |
2JCB
 
 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
