7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYC
 
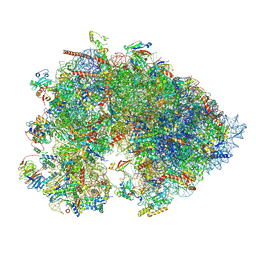 | | Cryo-EM structure of the Xenopus egg 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
6T6M
 
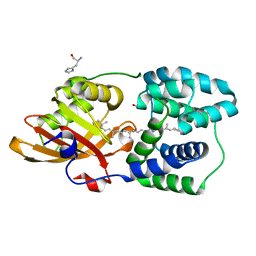 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
1A1O
 
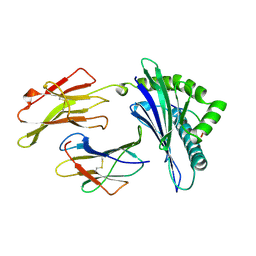 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
7EZM
 
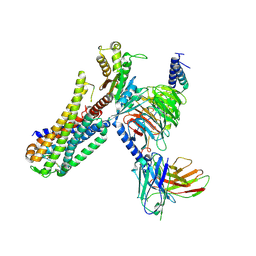 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZH
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
6Y30
 
 | | NG domain of human SRP54 T115A mutant | | Descriptor: | Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
8J47
 
 | | CryoEM Structure of 40-Residue Arctic (E22G) Beta-Amyloid Fibril Derived by Co-Analysis with Solid-State NMR | E22G Abeta40 | | Descriptor: | E22G Amyloid-beta | | Authors: | Tehrani, M.J, Matsuda, I, Yamagata, A, Matsunaga, T, Sato, M, Toyooka, K, Shirouzu, M, Ishii, Y, Kodama, Y, McElheny, D, Kobayashi, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | E22G A beta 40 fibril structure and kinetics illuminate how A beta 40 rather than A beta 42 triggers familial Alzheimer's.
Nat Commun, 15, 2024
|
|
6MNS
 
 | | Rhesus macaque anti-HIV V3 antibody DH753 with gp120 V3 ZAM18 peptide | | Descriptor: | Ab DH753 heavy chain Fab fragment, Ab DH753 light chain, Envelope glylcoprotein, ... | | Authors: | Nicely, N.I. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations.
Nat Commun, 10, 2019
|
|
6GEH
 
 | | Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. | | Descriptor: | DIMETHYL SULFOXIDE, FAD-binding 9, siderophore-interacting domain protein, ... | | Authors: | Trindade, I.B, Silva, J.P.M, Matias, P, Moe, E. | | Deposit date: | 2018-04-26 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and reactivity of a siderophore-interacting protein from the marine bacteriumShewanellareveals unanticipated functional versatility.
J. Biol. Chem., 294, 2019
|
|
7EZK
 
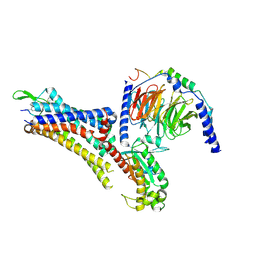 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex | | Descriptor: | Chimera of Guanine nucleotide-binding protein G(i) subunit alpha-1 and Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
2LDI
 
 | |
5VLI
 
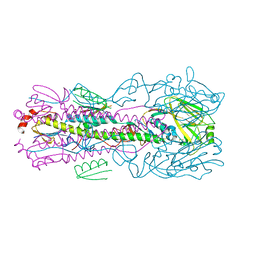 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
7W1G
 
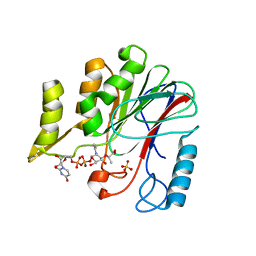 | | Crystal structure of YfiH with C107A mutation in complex with UDP-MurNAc-L-Serine | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, (2S)-2-[[(2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2-[[[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-6-(hydroxymethyl)-5-oxidanyl-oxan-4-yl]oxypropanoyl]amino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lee, S.H, Hsieh, K.Y, Lee, M.S, Chang, C.I. | | Deposit date: | 2021-11-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for the Peptidoglycan-Editing Activity of YfiH.
Mbio, 13, 2021
|
|
8TXT
 
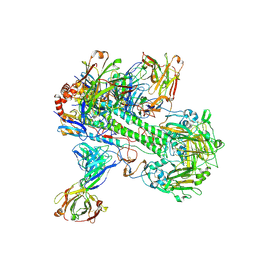 | | Crystal structure of 05.GC.w13.02 Fab in complex with H5 HA from A/Viet Nam/1203/2004(H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_W13B_B, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
5VXR
 
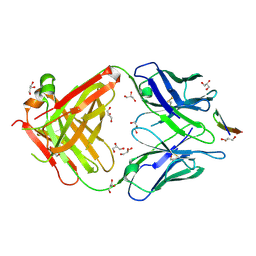 | | The antigen-binding fragment of MAb24 in complex with a peptide from Hepatitis C Virus E2 epitope I (412-423) | | Descriptor: | GLYCEROL, MAb24 Variable Heavy Chain,MAb24 Variable Heavy Chain, MAb24 Variable Light Chain,MAb24 Variable Light Chain, ... | | Authors: | Hardy, J.M, Gu, J, Boo, I, Drummer, H.E, Coulibaly, F.J. | | Deposit date: | 2017-05-24 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.
J. Virol., 92, 2018
|
|
6GGS
 
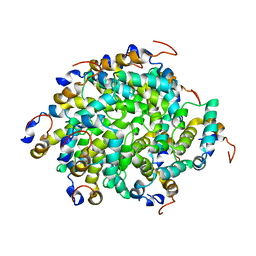 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
4RWS
 
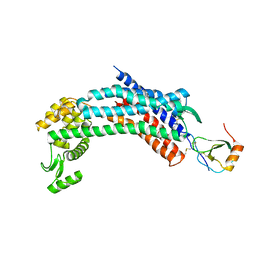 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
4ECM
 
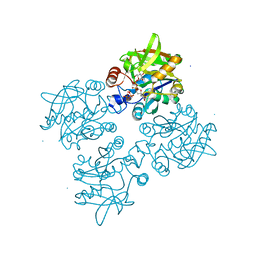 | | 2.3 Angstrom Crystal Structure of a Glucose-1-phosphate Thymidylyltransferase from Bacillus anthracis in Complex with Thymidine-5-diphospho-alpha-D-glucose and Pyrophosphate | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase, PYROPHOSPHATE 2- | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6GN2
 
 | | Racemic crystal structure of A-DNA duplex formed from d(CCCGGG) in space group R3 | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), trimethylamine oxide | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2018-05-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Racemic crystal structures of A-DNA duplexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8OJ8
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
4S1K
 
 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 100 K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
7NRB
 
 | | Re-refinement of MK3-inhibitor complex | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Croll, T.I, Read, R.J. | | Deposit date: | 2021-03-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptive Cartesian and torsional restraints for interactive model rebuilding.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3H9R
 
 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
6GHD
 
 | | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases | | Descriptor: | 1,2-ETHANEDIOL, Barrier-to-autointegration factor, Emerin, ... | | Authors: | Samson, C, Petitalot, A, Celli, F, Herrada, I, Ropars, V, Ledu, M.H, Nhiri, N, Arteni, A.A, Buendia, B, ZinnJustin, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases.
Nucleic Acids Res., 46, 2018
|
|
