5Y53
 
 | |
5YTX
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAACU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*AP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
6JKK
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
5YTT
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAUGU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*GP*U)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTV
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCAUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5ZNL
 
 | | Crystal structure of PDE10A catalytic domain complexed with LHB-6 | | Descriptor: | MAGNESIUM ION, N-[2-(7-methoxy-4-morpholin-4-yl-quinazolin-6-yl)oxyethyl]-1,3-benzothiazol-2-amine, ZINC ION, ... | | Authors: | Li, Z, Huang, Y, Zhan, C.G, Luo, H.B. | | Deposit date: | 2018-04-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Absolute Binding Free Energy Calculation and Design of a Subnanomolar Inhibitor of Phosphodiesterase-10.
J. Med. Chem., 62, 2019
|
|
6IRB
 
 | | C-terminal coiled coil domain of Drosophila phospholipase C beta NORPA, selenomethionine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
5XVJ
 
 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
6A6J
 
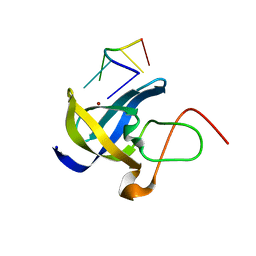 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
5Y21
 
 | |
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
7V6G
 
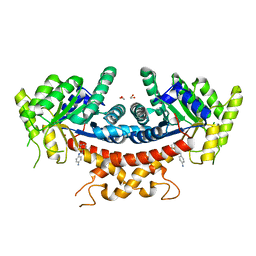 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
7V6F
 
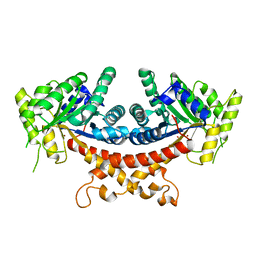 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
8H4R
 
 | | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, G1/S-specific cyclin-E1, ... | | Authors: | Gui, W, Wang, F, Cheng, W, Gao, J, Huang, Y, Ouyang, Z. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of CDK3 and CyclinE1 Complex with Dinaciclib from Biortus
To Be Published
|
|
8IA5
 
 | | Small peptide enhances the binding of nutline-3a to N-terminal domain of MdmX | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, 9-mer peptide, ... | | Authors: | Cheng, X.Y, Wei, Q.Y, Huang, Y, Su, Z.D. | | Deposit date: | 2023-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small peptide enhances the binding of nutline-3a to N-terminal domain of MdmX
To Be Published
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
5XYV
 
 | |
5Z9X
 
 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
5Z9Z
 
 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5ZUM
 
 | | Structure of dipeptidyl-peptidase III from Corallococcus sp. strain EGB | | Descriptor: | ZINC ION, dipeptidyl-peptidase III | | Authors: | Zhang, H, Duan, Y.J, Li, Z.K, Liu, W.D, Huang, Y, Cui, Z.L. | | Deposit date: | 2018-05-08 | | Release date: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of dipeptidyl peptidase III from Corallococcus sp. strain EGB
To Be Published
|
|
