5EW6
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5XG4
 
 | | Crystal structure of uPA in complex with quercetin | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural mechanism of flavonoids in inhibiting serine proteases
Food Funct, 8, 2017
|
|
3P8F
 
 | |
3PB1
 
 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
3P8G
 
 | | Crystal Structure of MT-SP1 in complex with benzamidine | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, GLUTATHIONE, ... | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of catalytic domain of Matriptase in complex with Sunflower trypsin inhibitor-1.
Bmc Struct.Biol., 11, 2011
|
|
4DVB
 
 | | The crystal structure of the Fab fragment of pro-uPA antibody mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
4DVA
 
 | | The crystal structure of human urokinase-type plasminogen activator catalytic domain | | Descriptor: | HEXAETHYLENE GLYCOL, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody
Biochem.J., 449, 2013
|
|
4DW2
 
 | | The crystal structure of uPA in complex with the Fab fragment of mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
3S8B
 
 | | Structure of Yeast Ribonucleotide Reductase 1 with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S8A
 
 | | Structure of Yeast Ribonucleotide Reductase R293A with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S87
 
 | | Structure of Yeast Ribonucleotide Reductase 1 with dGTP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3S8C
 
 | | Structure of Yeast Ribonucleotide Reductase 1 R293A with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
6KA4
 
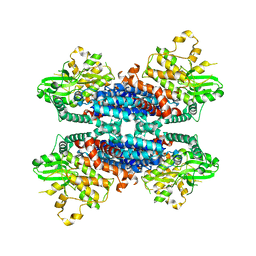 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
3JB9
 
 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCM
 
 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
4OJV
 
 | | Crystal structure of unliganded yeast PDE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, SULFATE ION, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
4QGE
 
 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | Descriptor: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | Authors: | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | Deposit date: | 2014-05-22 | | Release date: | 2014-12-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
4ZHM
 
 | |
4ZKO
 
 | |
4ZKS
 
 | |
4ZKR
 
 | |
4ZKN
 
 | |
