7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
5YVX
 
 | | Crystal structure of SDG8 CW domain in complex with H3K4me1 peptide | | Descriptor: | H3K4me1, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Liu, Y, Huang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain ofArabidopsishistone methyltransferase SDG8.
J. Biol. Chem., 293, 2018
|
|
2QC3
 
 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
5XVW
 
 | |
5XVL
 
 | | Crystal structure of AL2 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 2, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
5Y21
 
 | |
5Y53
 
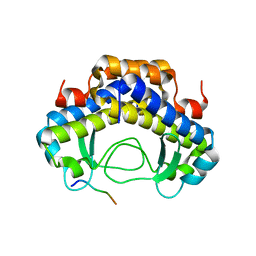 | |
5YTX
 
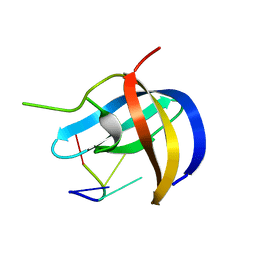 | | Crystal structure of YB1 cold-shock domain in complex with UCAACU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*AP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTT
 
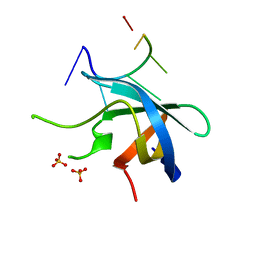 | | Crystal structure of YB1 cold-shock domain in complex with UCAUGU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*GP*U)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
5YTV
 
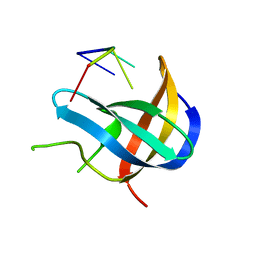 | | Crystal structure of YB1 cold-shock domain in complex with UCAUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*UP*CP*AP*UP*CP*U)-3') | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
8FXF
 
 | | Crystal structure of the coiled-coil domain of TRIM56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase TRIM56 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.
Comput Struct Biotechnol J, 21, 2023
|
|
5ZNL
 
 | | Crystal structure of PDE10A catalytic domain complexed with LHB-6 | | Descriptor: | MAGNESIUM ION, N-[2-(7-methoxy-4-morpholin-4-yl-quinazolin-6-yl)oxyethyl]-1,3-benzothiazol-2-amine, ZINC ION, ... | | Authors: | Li, Z, Huang, Y, Zhan, C.G, Luo, H.B. | | Deposit date: | 2018-04-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Absolute Binding Free Energy Calculation and Design of a Subnanomolar Inhibitor of Phosphodiesterase-10.
J. Med. Chem., 62, 2019
|
|
5XVJ
 
 | | Crystal structure of AL7 PAL domain | | Descriptor: | PHD finger protein ALFIN-LIKE 7, SULFATE ION | | Authors: | Peng, L, Wang, L.L, Huang, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Arabidopsis AL2-PAL and PRC1 Complex Provides Mechanistic Insight into Active-to-Repressive Chromatin State Switch
J. Mol. Biol., 430, 2018
|
|
6A6J
 
 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7V6G
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase mutation C157S with CN39 | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION, ... | | Authors: | Cao, H, Huang, Y, Chen, H, Wan, C, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
7V6F
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
3TJ2
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[(4-chlorobenzyl)(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, POTASSIUM ION | | Authors: | Wolf, S, Huang, Y, Popowicz, G.M, Goda, S, Holak, T.A, Doemling, A. | | Deposit date: | 2011-08-23 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ugi Multicomponent Reaction Derived p53-Mdm2 Antagonists
To be published
|
|
3TU1
 
 | | Exhaustive Fluorine Scanning towards Potent p53-MDM2 Antagonist | | Descriptor: | 3-[(1S)-2-(tert-butylamino)-1-{N-[(3,4-difluorophenyl)methyl]formamido}-2-oxoethyl]-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wolf, S, Huang, Y, Koes, D, Popowicz, G.M, Camacho, C.J, Holak, T.A, Doemling, A. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Exhaustive Fluorine Scanning toward Potent p53-Mdm2 Antagonists.
Chemmedchem, 7, 2012
|
|
7JST
 
 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT0
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT7
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
3TG2
 
 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | Authors: | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TGV
 
 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
