3WQK
 
 | | Crystal structure of Rv3378c with PO4 | | Descriptor: | Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3W02
 
 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3WQM
 
 | | Crystal structure of Rv3378c with inhibitor BPH-629 | | Descriptor: | Diterpene synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQN
 
 | | Crystal structure of Rv3378c_Y51F with TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQL
 
 | | Crystal structure of Rv3378c with Mg2+ and PPi | | Descriptor: | Diterpene synthase, MAGNESIUM ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3VNC
 
 | | Crystal Structure of TIP-alpha N25 from Helicobacter Pylori in its natural dimeric form | | Descriptor: | TIP-alpha | | Authors: | Gao, M, Li, D, Hu, Y, Zou, Q, Wang, D.-C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TNF-alpha-Inducing Protein from Helicobacter Pylori in Active Form Reveals the Intrinsic Molecular Flexibility for Unique DNA-Binding
Plos One, 7, 2012
|
|
7JMB
 
 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | Authors: | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7JMA
 
 | |
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
3PDI
 
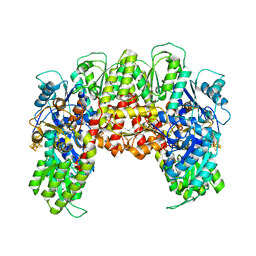 | | Precursor bound NifEN | | Descriptor: | IRON/SULFUR CLUSTER, L-Cluster (Fe8S9), Nitrogenase MoFe cofactor biosynthesis protein NifE, ... | | Authors: | Kaiser, J.T, Hu, Y, Wiig, J.A, Rees, D.C, Ribbe, M.W. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Science, 331, 2011
|
|
3QTF
 
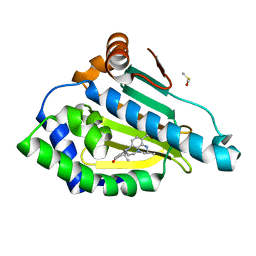 | | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity | | Descriptor: | (6S)-6,15,15,18-tetramethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-8,12-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Ingalls, C, Sutherland, A.G, Sonye, J.P, Eid, C.N, Golas, J, Liu, H, Boschelli, F, Hu, Y, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5703 Å) | | Cite: | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R91
 
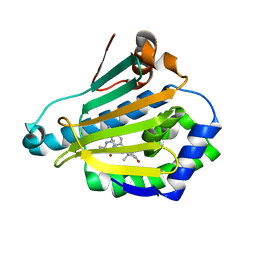 | | Macrocyclic lactams as potent Hsp90 inhibitors with excellent tumor exposure and extended biomarker activity. | | Descriptor: | (6S)-4,6,15,15,18-pentamethyl-5,17-dioxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Otteng, M, Ingalls, C, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Hu, Y, Levin, J.I. | | Deposit date: | 2011-03-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Macrocyclic lactams as potent Hsp90 inhibitors with excellent tumor exposure and extended biomarker activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8GQ6
 
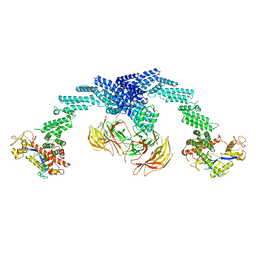 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
4G3H
 
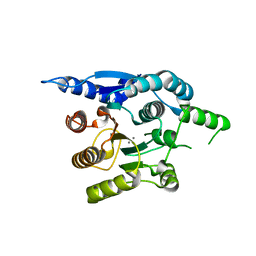 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
2M6S
 
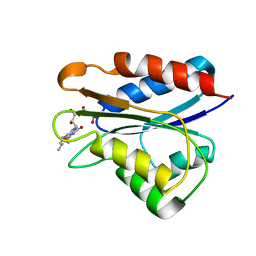 | | Holo_YqcA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2P0Q
 
 | |
2MOK
 
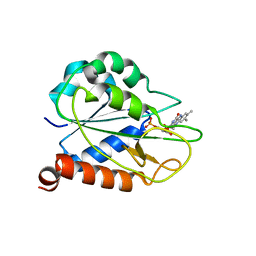 | | holo_FldA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2014-04-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR study of YqcA from Escherichia coli
To be Published
|
|
2P0P
 
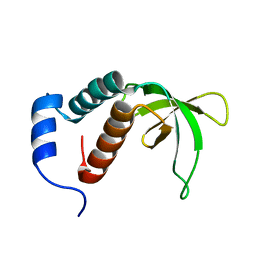 | | Calcium binding protein in the free form | | Descriptor: | Alr1010 protein | | Authors: | Zhang, X, Hu, Y, Jin, C. | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Ccbp from Anabaena Reveals a New Fold and Novel Calcium Binding Sites
To be Published
|
|
2M6R
 
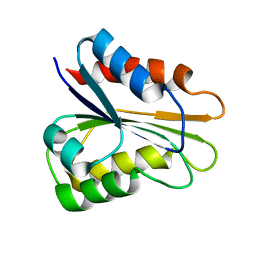 | | apo_YqcA | | Descriptor: | Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2MYJ
 
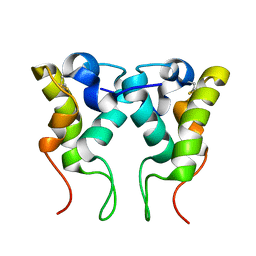 | |
7W1W
 
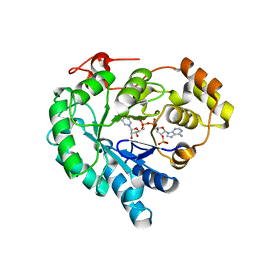 | | NADPH-bound AKR4C17 mutant F291D | | Descriptor: | AKR4-2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7W1X
 
 | | Crystal structure of AKR4C16 bound with NADPH | | Descriptor: | AKR4-1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
