6M4H
 
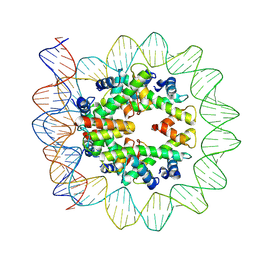 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (103-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
9AYM
 
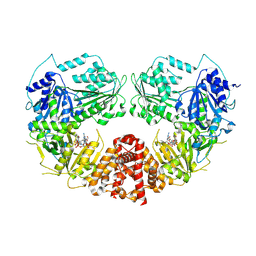 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9B0I
 
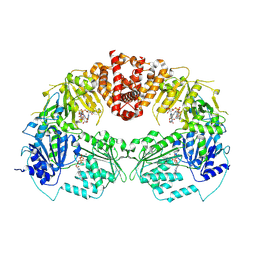 | |
3KT3
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
6XCG
 
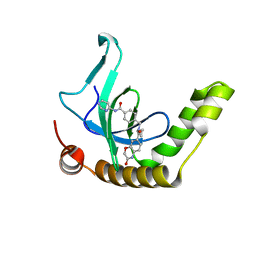 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
6XKB
 
 | | Crystal structure of SR-related and CTD-associated factor 4(SCAF4-CID)with peptide S2,S5p-CTD | | Descriptor: | S2,S5p-CTD peptide, SR-related and CTD-associated factor 4, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Febs Lett., 596, 2022
|
|
6M4D
 
 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (125-MER), Histone H2A.V, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6V4L
 
 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6V4J
 
 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6V4K
 
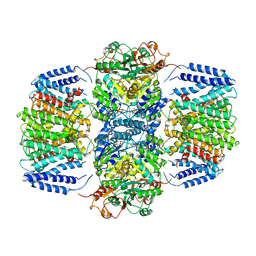 | | Structure of TrkH-TrkA in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Potassium transporter peripheral membrane component, Trk system potassium uptake protein | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.53004146 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
9B0E
 
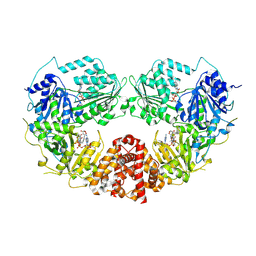 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-amide-CoA bisubstrate probe and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA cytidine acetyltransferase, [[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[[1-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-2-oxidanylidene-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
7UVE
 
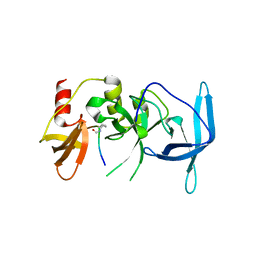 | | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac | | Descriptor: | Histone-lysine N-methyltransferase eggless, peptide H3K9me2K14ac | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac
To Be Published
|
|
7UW8
 
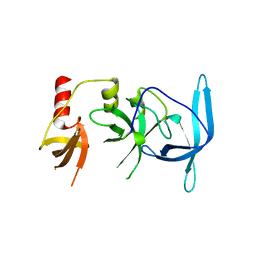 | | Drosophila melanogaster setdb1-tuor domain | | Descriptor: | Histone-lysine N-methyltransferase eggless | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain
To Be Published
|
|
3KT6
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with Trp | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KT0
 
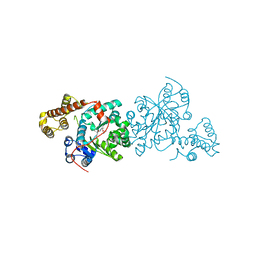 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase | | Descriptor: | (2S)-1-{[(2S)-3-(2-methoxyethoxy)-2-methylpropyl]oxy}propan-2-amine, Tryptophanyl-tRNA synthetase, cytoplasmic | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3KT8
 
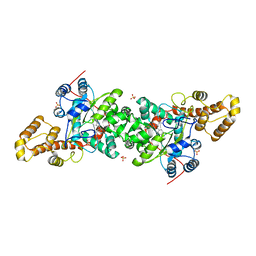 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3PPX
 
 | | Crystal structure of the N1602A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPW
 
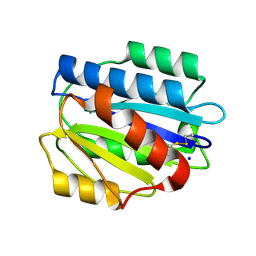 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
3PPY
 
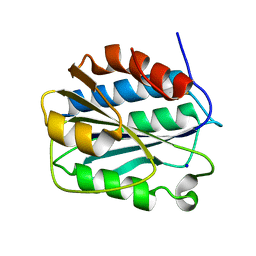 | | Crystal structure of the D1596A/N1602A double mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
5NI1
 
 | | CryoEM structure of haemoglobin at 3.2 A determined with the Volta phase plate | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Khoshouei, M, Radjainia, M, Bunker, R, Baumeister, W, Danev, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of haemoglobin at 3.2 angstrom determined with the Volta phase plate.
Nat Commun, 8, 2017
|
|
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | Authors: | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
1TCE
 
 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
5YYN
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate TRNA(Arg) | | Descriptor: | Arginine--tRNA ligase, TRNA | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
5YYM
 
 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate Arg | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
