4UNT
 
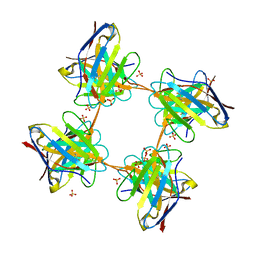 | | Induced monomer of the Mcg variable domain | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C.M, Whitelegge, J.P, Phillips, M.L, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
5TVO
 
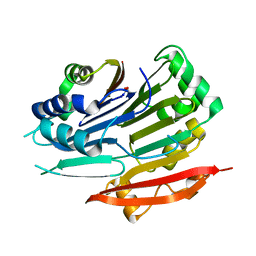 | | Crystal structure of Trypanosoma brucei AdoMetDC-delta26 monomer | | Descriptor: | S-adenosylmethionine decarboxylase alpha chain, S-adenosylmethionine decarboxylase proenzyme, SODIUM ION | | Authors: | Volkov, O.A, Ariagno, C, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
4QR9
 
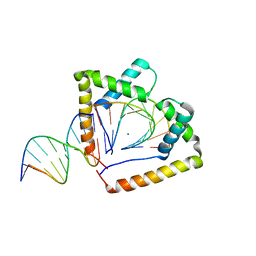 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | Authors: | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6DFT
 
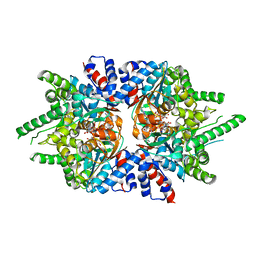 | | Trypanosoma brucei deoxyhypusine synthase | | Descriptor: | Deoxyhypusine synthase, Deoxyhypusine synthase regulatory subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tomchick, D.R, Phillips, M.A, Afanador, G.A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trypanosomatid Deoxyhypusine Synthase Activity Is Dependent on Shared Active-Site Complementation between Pseudoenzyme Paralogs.
Structure, 26, 2018
|
|
6BM7
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer in complex with pyrimidineamine inhibitor UTSAM568 | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-4-[(3,5-dibromophenyl)amino]-6-methylpyrimidin-1-ium, ... | | Authors: | Volkov, O.A, Chen, Z, Phillips, M.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Species-Selective Pyrimidineamine Inhibitors of Trypanosoma brucei S-Adenosylmethionine Decarboxylase.
J. Med. Chem., 61, 2018
|
|
5TVM
 
 | | Crystal structure of Trypanosoma brucei AdoMetDC/prozyme heterodimer | | Descriptor: | 1,4-DIAMINOBUTANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Volkov, O.A, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
5ACM
 
 | | Mcg immunoglobulin variable domain with methylene blue | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, GLYCEROL, MCG, ... | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
6E0B
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase C276F mutant bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Tomchick, D.R, Phillips, M.A, Deng, X. | | Deposit date: | 2018-07-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification and Mechanistic Understanding of Dihydroorotate Dehydrogenase Point Mutations in Plasmodium falciparum that Confer in Vitro Resistance to the Clinical Candidate DSM265.
ACS Infect Dis, 5, 2019
|
|
5ACL
 
 | | Mcg immunoglobulin variable domain with sulfasalazine | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, MCG, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3QH9
 
 | | Human Liprin-beta2 Coiled-Coil | | Descriptor: | AMMONIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Stafford, R.L, Tang, M, Phillips, M.L, Bowie, J.U. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the central coiled-coil domain from human liprin-beta2
Biochemistry, 50, 2011
|
|
5TBO
 
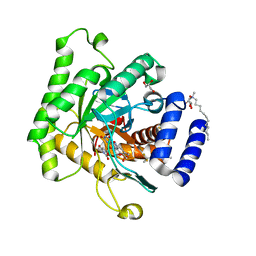 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM421 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[6-(trifluoromethyl)pyridin-3-yl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | A Triazolopyrimidine-Based Dihydroorotate Dehydrogenase Inhibitor with Improved Drug-like Properties for Treatment and Prevention of Malaria.
ACS Infect Dis, 2, 2016
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3MAZ
 
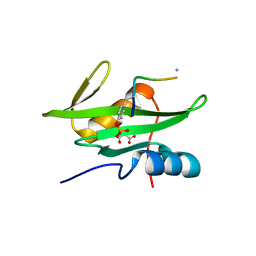 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
3PLS
 
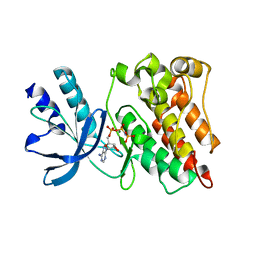 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
1JI7
 
 | | Crystal Structure of TEL SAM Polymer | | Descriptor: | ETS-RELATED PROTEIN TEL1, SULFATE ION | | Authors: | Kim, C.A, Phillips, M.L, Kim, W, Gingery, M, Tran, H.H, Robinson, M.A, Faham, S, Bowie, J.U. | | Deposit date: | 2001-06-29 | | Release date: | 2002-07-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression.
EMBO J., 20, 2001
|
|
3FGH
 
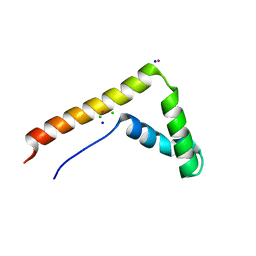 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
3ZEV
 
 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HMA
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
3I6R
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM74 | | Descriptor: | 5-methyl-N-[4-(trifluoromethyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
3I65
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
3I68
 
 | |
4BV0
 
 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BUO
 
 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BWB
 
 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
