3DYA
 
 | | HIV-1 RT with non-nucleoside inhibitor annulated Pyrazole 1 | | Descriptor: | 3-[6-bromo-2-fluoro-3-(1H-pyrazolo[3,4-c]pyridazin-3-ylmethyl)phenoxy]-5-chlorobenzonitrile, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-07-25 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of annulated pyrazoles as inhibitors of HIV-1 reverse transcriptase
J.Med.Chem., 51, 2008
|
|
7T98
 
 | |
7T97
 
 | |
6EK2
 
 | |
6EJG
 
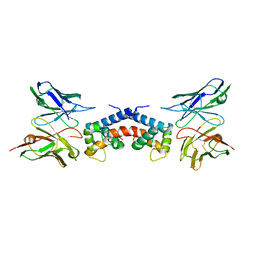 | |
2GFS
 
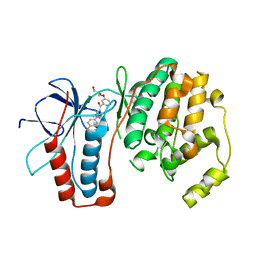 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
2GIR
 
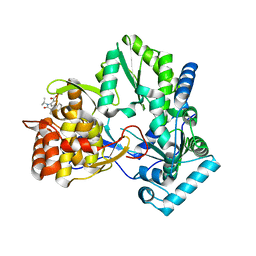 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-1 inhibitor | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GIQ
 
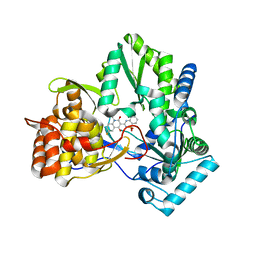 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-2 inhibitor | | Descriptor: | 1-(2-CYCLOPROPYLETHYL)-3-(1,1-DIOXIDO-2H-1,2,4-BENZOTHIADIAZIN-3-YL)-6-FLUORO-4-HYDROXYQUINOLIN-2(1H)-ONE, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GQ0
 
 | |
3NBP
 
 | | HIV-1 reverse transcriptase with aminopyrimidine inhibitor 2 | | Descriptor: | 4-(4-{[4-(4-cyano-2,6-dimethylphenoxy)pyrimidin-2-yl]amino}piperidin-1-yl)benzenesulfonamide, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1QQH
 
 | |
1SF8
 
 | | Crystal structure of the carboxy-terminal domain of htpG, the E. coli Hsp90 | | Descriptor: | CHLORIDE ION, Chaperone protein htpG, NICKEL (II) ION | | Authors: | Harris, S.F, Shiau, A.K, Agard, D.A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site.
Structure, 12, 2004
|
|
4ZFF
 
 | | Dual-acting Fab 5A12 in complex with VEGF | | Descriptor: | Fragment antigen binding (Fab) 5A12 Heavy chain, Fragment antigen binding (Fab) 5A12 Light chain, SULFATE ION, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration.
J.Biol.Chem., 290, 2015
|
|
4ZK5
 
 | | MAP4K4 in complex with inhibitor GNE-495 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-amino-N-[1-(cyclopropylcarbonyl)azetidin-3-yl]-2-(3-fluorophenyl)-1,7-naphthyridine-5-carboxamide, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2015-04-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis.
Acs Med.Chem.Lett., 6, 2015
|
|
4ZFG
 
 | | Dual-specificity Fab 5A12 in complex with Angiopoietin 2 | | Descriptor: | Angiopoietin-2, CALCIUM ION, Fragment antigen binding 5A12 heavy chain, ... | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration.
J.Biol.Chem., 290, 2015
|
|
5CEO
 
 | | DLK in complex with inhibitor 2-((6-(3,3-difluoropyrrolidin-1-yl)-4-(1-(oxetan-3-yl)piperidin-4-yl)pyridin-2-yl)amino)isonicotinonitrile | | Descriptor: | 2-[[6-[3,3-bis(fluoranyl)pyrrolidin-1-yl]-4-[1-(oxetan-3-yl)piperidin-4-yl]pyridin-2-yl]amino]pyridine-4-carbonitrile, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CEN
 
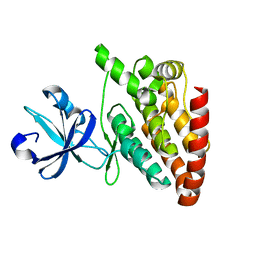 | | Crystal structure of DLK (kinase domain) | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CEP
 
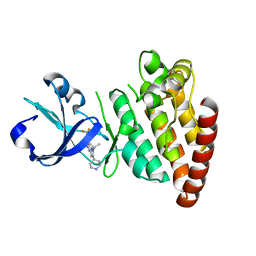 | |
5CEQ
 
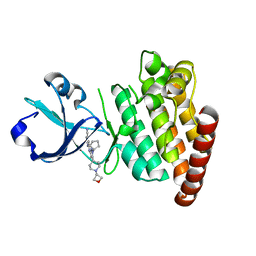 | |
5CVM
 
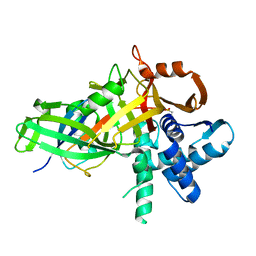 | | USP46~ubiquitin BEA covalent complex | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 46, ZINC ION | | Authors: | Harris, S.F, Yin, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
5CVL
 
 | | WDR48 (UAF-1), residues 2-580 | | Descriptor: | GOLD ION, PHOSPHATE ION, WD repeat-containing protein 48 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into WD-Repeat 48 Activation of Ubiquitin-Specific Protease 46.
Structure, 23, 2015
|
|
3M8P
 
 | | HIV-1 RT with NNRTI TMC-125 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2010-03-18 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3M8Q
 
 | | HIV-1 RT with AMINOPYRIMIDINE NNRTI | | Descriptor: | 3,5-dimethyl-4-{[2-({1-[4-(methylsulfonyl)benzyl]piperidin-4-yl}amino)pyrimidin-4-yl]oxy}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2010-03-18 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MF5
 
 | |
3H5S
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor | | Descriptor: | (5S)-5-tert-butyl-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-3-[8-(methylsulfonyl)-1,1-dioxido-6,7,8,9-tetrahydroisothiazolo[4,5-h]isoquinolin-3-yl]-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
