3C1C
 
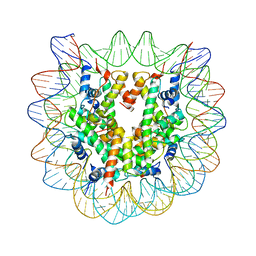 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
To be published
|
|
8P2Y
 
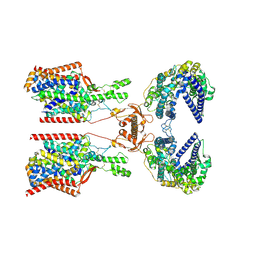 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
To be published
|
|
8P30
 
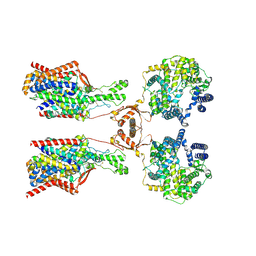 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
TO BE CONFIRMED
|
|
8P31
 
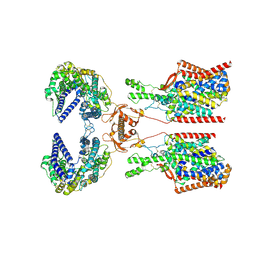 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
To Be Published
|
|
8P2W
 
 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
TO BE CONFIRMED
|
|
8P2Z
 
 | | Structure of human SIT1 bound to L-pipecolate (focussed map / refinement) | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2
TO BE CONFIRMED
|
|
8BIK
 
 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
4H1W
 
 | | E1 structure of the (SR) Ca2+-ATPase in complex with Sarcolipin | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bublitz, M, Winther, A.-M.L, Karlsen, J, Moller, J.V, Hansen, J.B, Buch-Pedersen, M.J, Nissen, P. | | Deposit date: | 2012-09-11 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm.
Nature, 495, 2013
|
|
1FFK
 
 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
1FFZ
 
 | | LARGE RIBOSOMAL SUBUNIT COMPLEXED WITH R(CC)-DA-PUROMYCIN | | Descriptor: | 23S RIBOSOMAL RNA, R(P*CP*C*)-D(P*A)-R(P*(PU)) | | Authors: | Nissen, P, Hansen, J, Ban, N, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of ribosome activity in peptide bond synthesis.
Science, 289, 2000
|
|
1FG0
 
 | | LARGE RIBOSOMAL SUBUNIT COMPLEXED WITH A 13 BP MINIHELIX-PUROMYCIN COMPOUND | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(CCGGCGGGCUGGUUCAAACCGGCCCGCCGGACC)-3'-5'-R(P-PUROMYCIN)-3' | | Authors: | Nissen, P, Hansen, J, Ban, N, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of ribosome activity in peptide bond synthesis.
Science, 289, 2000
|
|
4PP7
 
 | | Highly Potent and Selective 3-N-methylquinazoline-4(3H)-one Based Inhibitors of B-RafV600E Kinase | | Descriptor: | N-{2,4-difluoro-3-[methyl(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wenglowsky, S, Ren, L, Grina, J, Hansen, J.D, Laird, E.R, Moreno, D, Dinkel, V, Gloor, S.L, Hastings, G, Rana, S, Rasor, K, Sturgis, H.L, Voegtli, W.C, Vigers, G.P.A, Willis, B, Mathieu, S, Rudolph, J. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Highly potent and selective 3-N-methylquinazoline-4(3H)-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7JZV
 
 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1EJ8
 
 | | CRYSTAL STRUCTURE OF DOMAIN 2 OF THE YEAST COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (LYS7) AT 1.55 A RESOLUTION | | Descriptor: | CALCIUM ION, LYS7 | | Authors: | Hall, L.T, Sanchez, R.J, Holloway, S.P, Zhu, H, Stine, J.E, Lyons, T.J, Demeler, B, Schirf, V, Hansen, J.C, Nersissian, A.M, Valentine, J.S, Hart, P.J. | | Deposit date: | 2000-03-01 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): a dimer-dimer model of LYS7-SOD association and copper delivery.
Biochemistry, 39, 2000
|
|
8P4K
 
 | | Vaccinia Virus palisade layer A10 trimer | | Descriptor: | Core protein OPG136 | | Authors: | Datler, J, Hansen, J.M, Thader, A, Schloegl, A, Hodirnau, V.V, Schur, F.K.M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Multi-modal cryo-EM reveals trimers of protein A10 to form the palisade layer in poxvirus cores.
Nat.Struct.Mol.Biol., 2024
|
|
5NCQ
 
 | | Structure of the (SR) Ca2+-ATPase bound to a Tetrahydrocarbazole and TNP-ATP | | Descriptor: | (1~{S})-~{N}-[(4-bromophenyl)methyl]-7-(trifluoromethyloxy)-2,3,4,9-tetrahydro-1~{H}-carbazol-1-amine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, ... | | Authors: | Bublitz, M, Kjellerup, L, O'Hanlon Cohrt, K, Gordon, S, Mortensen, A.L, Clausen, J.D, Pallin, D, Hansen, J.B, Brown, W.D, Fuglsang, A, Winther, A.-M.L. | | Deposit date: | 2017-03-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity.
PLoS ONE, 13, 2018
|
|
1KQS
 
 | | The Haloarcula marismortui 50S Complexed with a Pretranslocational Intermediate in Protein Synthesis | | Descriptor: | 23S RRNA, 5S RRNA, 6-AMINOHEXANOIC ACID, ... | | Authors: | Schmeing, T.M, Seila, A.C, Hansen, J.L, Freeborn, B, Soukup, J.K, Scaringe, S.A, Strobel, S.A, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-01-07 | | Release date: | 2002-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits.
Nat.Struct.Biol., 9, 2002
|
|
