6Y4S
 
 | |
6SJU
 
 | |
6SHI
 
 | |
6SHH
 
 | |
5KW9
 
 | |
5WPI
 
 | |
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P26
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with A-type 2 HBGA | | Descriptor: | P domain of VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P1V
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
3SEJ
 
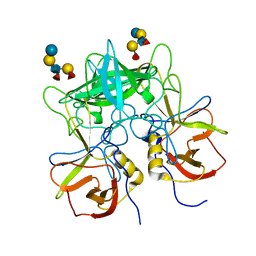 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to Secretor Lewis HBGA (LeB) | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLD
 
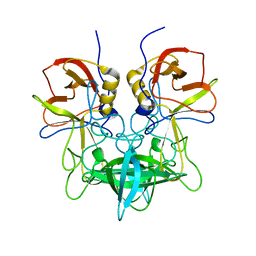 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to A trisaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SLN
 
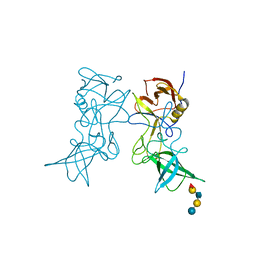 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SKB
 
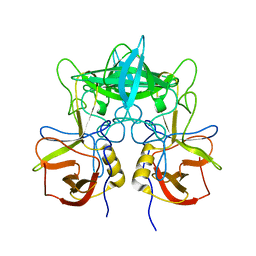 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3SJP
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) | | Descriptor: | Capsid, ZINC ION | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
4FKD
 
 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, CITRATE ANION, ZINC ION | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
8HAE
 
 | | Cryo-EM structure of HACE1 dimer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8H8X
 
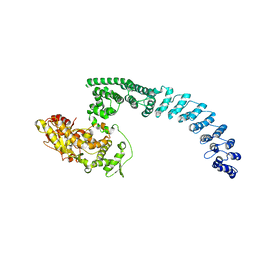 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
3JYI
 
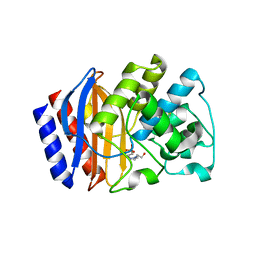 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
7LUP
 
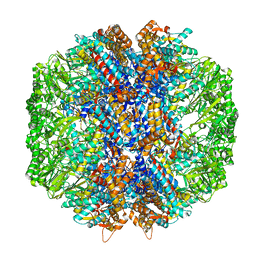 | | Human TRiC/CCT complex with reovirus outer capsid protein sigma-3 | | Descriptor: | Outer capsid protein sigma-3, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LUM
 
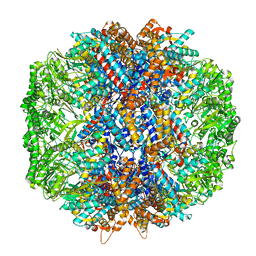 | | Human TRiC in ATP/AlFx closed state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4F6H
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
