7VQO
 
 | | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | | Descriptor: | Ams1, Nbr1 and malE fusion protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1
Nat Commun, 13, 2022
|
|
6VZO
 
 | |
6VZM
 
 | |
6VZN
 
 | |
6VZL
 
 | |
6AVI
 
 | |
6AUG
 
 | |
6O67
 
 | |
6O68
 
 | |
6DHA
 
 | |
7DDE
 
 | | Cryo-EM structure of the Ape4 and Nbr1 complex | | Descriptor: | Aspartyl aminopeptidase 1,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
7DD9
 
 | | Cryo-EM structure of the Ams1 and Nbr1 complex | | Descriptor: | Alpha-mannosidase,ZZ-type zinc finger-containing protein P35G2.11c,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2020-10-28 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1.
Embo J., 40, 2021
|
|
6C1I
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
8WYF
 
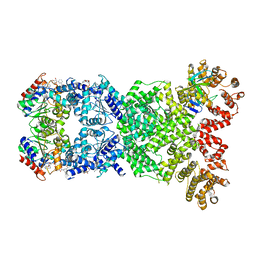 | | Cryo-EM structure of DSR2-DSAD1-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8XG2
 
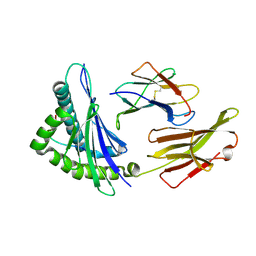 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8WY8
 
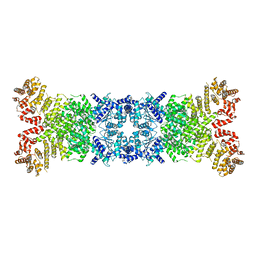 | | Cryo-EM structure of DSR2 apo complex | | Descriptor: | SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8XES
 
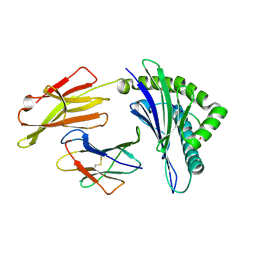 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
6AIX
 
 | |
6AIY
 
 | |
7WKJ
 
 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
6ONJ
 
 | |
6ONI
 
 | |
7XG3
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG0
 
 | | CryoEM structure of type IV-A Csf-crRNA-dsDNA ternary complex | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
7XG4
 
 | | CryoEM structure of type IV-A CasDinG bound NTS-nicked Csf-crRNA-dsDNA quaternary complex in a second state | | Descriptor: | Csf1, Csf2, Csf3, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-04-02 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
