1I1E
 
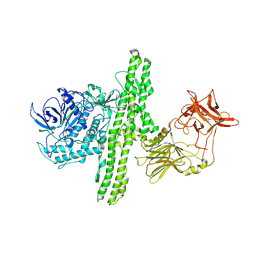 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM BOTULINUM NEUROTOXIN B COMPLEXED WITH DOXORUBICIN | | 分子名称: | BOTULINUM NEUROTOXIN TYPE B, DOXORUBICIN, SULFATE ION, ... | | 著者 | Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | 登録日 | 2001-02-01 | | 公開日 | 2001-11-21 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystallographic evidence for doxorubicin binding to the receptor-binding site in Clostridium botulinum neurotoxin B.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7CGB
 
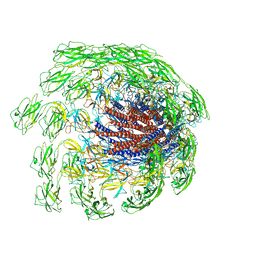 | | Cryo-EM structure of the flagellar hook from Salmonella | | 分子名称: | Flagellar hook protein FlgE | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-07-01 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | 分子名称: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | 分子名称: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
4OO9
 
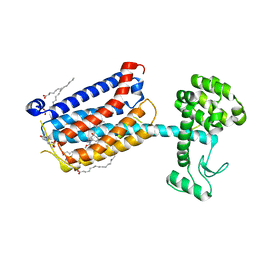 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | 著者 | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | 登録日 | 2014-01-31 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
7NOW
 
 | |
2XXA
 
 | | The Crystal Structure of the Signal Recognition Particle (SRP) in Complex with its Receptor(SR) | | 分子名称: | 4.5S RNA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | 著者 | Ataide, S.F, Schmitz, N, Shen, K, Ke, A, Shan, S, Doudna, J.A, Ban, N. | | 登録日 | 2010-11-09 | | 公開日 | 2011-03-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.94 Å) | | 主引用文献 | The Crystal Structure of the Signal Recognition Particle in Complex with its Receptor.
Science, 331, 2011
|
|
3UN1
 
 | | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021 | | 分子名称: | PHOSPHATE ION, Probable oxidoreductase | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-11-15 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021
To be Published
|
|
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | 分子名称: | hypothetical protein | | 著者 | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-02-09 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
1I4P
 
 | |
1I4R
 
 | |
1I4Q
 
 | |
1HSL
 
 | |
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | 登録日 | 2021-10-08 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | 登録日 | 2021-10-13 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7E
 
 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | 著者 | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2013-11-22 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
4V4I
 
 | | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements. | | 分子名称: | 16S SMALL SUBUNIT RIBOSOMAL RNA, 23S LARGE SUBUNIT RIBOSOMAL RNA, 30S ribosomal protein S10, ... | | 著者 | Korostelev, A, Trakhanov, S, Laurberg, M, Noller, H.F. | | 登録日 | 2007-02-15 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.71 Å) | | 主引用文献 | Crystal Structure of a 70S Ribosome-tRNA Complex Reveals Functional Interactions and Rearrangements
Cell(Cambridge,Mass.), 126, 2006
|
|
8GDU
 
 | | Crystal structure of a mutant methyl transferase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium (NESG) Target MvR53-11M | | 分子名称: | Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | 登録日 | 2023-03-06 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8HAE
 
 | | Cryo-EM structure of HACE1 dimer | | 分子名称: | E3 ubiquitin-protein ligase HACE1 | | 著者 | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | 登録日 | 2022-10-26 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (4.55 Å) | | 主引用文献 | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
3KZB
 
 | |
3L0Q
 
 | |
4V85
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | 登録日 | 2011-06-13 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
8H8X
 
 | | Cryo-EM structure of HACE1 monomer | | 分子名称: | E3 ubiquitin-protein ligase HACE1 | | 著者 | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | 登録日 | 2022-10-24 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
2G59
 
 | |
