6VUA
 
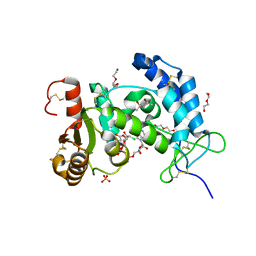 | | X-ray structure of human CD38 catalytic domain with 2'-Cl-araNAD+ | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Dai, Z, Zhang, X.N, Nasertorabi, F, Han, G.W, Stevens, R.C, Zhang, Y. | | 登録日 | 2020-02-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Synthesis of site-specific antibody-drug conjugates by ADP-ribosyl cyclases.
Sci Adv, 6, 2020
|
|
3ZKJ
 
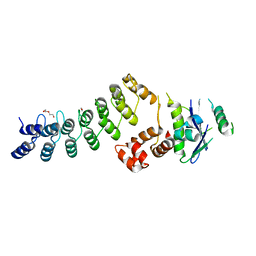 | | Crystal Structure of Ankyrin Repeat and Socs Box-Containing Protein 9 (Asb9) in Complex with Elonginb and Elonginc | | 分子名称: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, CHLORIDE ION, ... | | 著者 | Muniz, J.R.C, Guo, K, Zhang, Y, Ayinampudi, V, Savitsky, P, Keates, T, Filippakopoulos, P, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, von Delft, F, Knapp, S, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2013-01-23 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
8JCH
 
 | |
8JOL
 
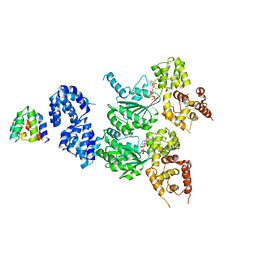 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | 著者 | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | 登録日 | 2023-06-07 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | 分子名称: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | 登録日 | 2022-10-09 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ISS
 
 | | Cryo-EM structure of wild-type human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG at 3.19 A resolution | | 分子名称: | MAGNESIUM ION, RNA (88-MER), tRNA-splicing endonuclease subunit Sen15, ... | | 著者 | Sun, Y, Zhang, Y, Yuan, L, Han, Y. | | 登録日 | 2023-03-21 | | 公開日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex.
Nat Commun, 14, 2023
|
|
3BH0
 
 | | ATPase Domain of G40P | | 分子名称: | DNAB-LIKE REPLICATIVE HELICASE | | 著者 | Wang, G, Klein, M.G, Tokonzaba, E, Zhang, Y, Holden, L.G, Chen, X.S. | | 登録日 | 2007-11-27 | | 公開日 | 2007-12-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The structure of a DnaB-family replicative helicase and its interactions with primase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1ZWZ
 
 | |
2Q5A
 
 | | human Pin1 bound to L-PEPTIDE | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Noel, J.P, Zhang, Y. | | 登録日 | 2007-05-31 | | 公開日 | 2007-06-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
3KEX
 
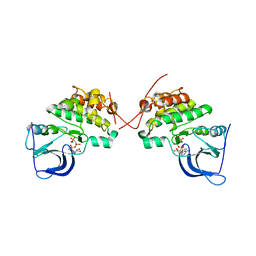 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | 著者 | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | 登録日 | 2009-10-26 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1R0V
 
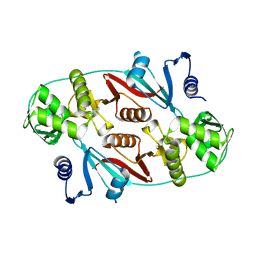 | |
1R11
 
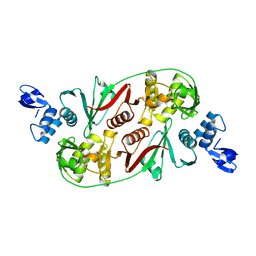 | |
4AOJ
 
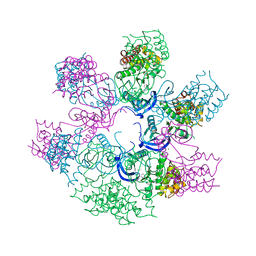 | | Human TrkA in complex with the inhibitor AZ-23 | | 分子名称: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | 著者 | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | 登録日 | 2012-03-28 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
7XHE
 
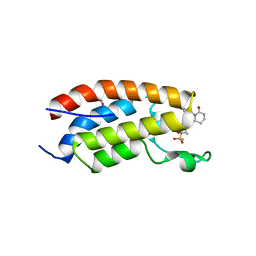 | | Crystal structure of CBP bromodomain liganded with CCS151 | | 分子名称: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-08 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XH6
 
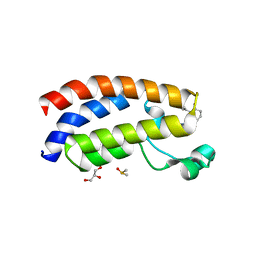 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | 分子名称: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
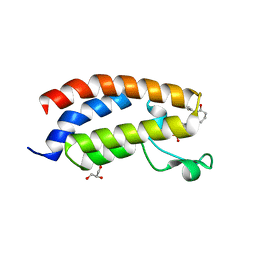 | | Crystal structure of CBP bromodomain liganded with CCS150 | | 分子名称: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-11 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | 分子名称: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-06 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | 分子名称: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | 著者 | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8BWU
 
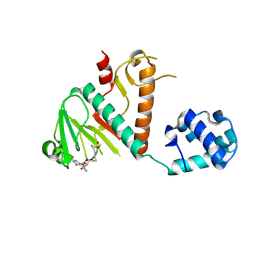 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | 著者 | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | 登録日 | 2022-12-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2022-04-21 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2022-04-21 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
