7E1B
 
 | | Crystal structure of VbrR-DNA complex | | Descriptor: | DNA (26-MER), DNA-binding response regulator | | Authors: | Hong, S, Zhang, X, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (4.587 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | Descriptor: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
7EGK
 
 | | Bicarbonate transporter complex SbtA-SbtB bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EGL
 
 | | Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | | Descriptor: | BICARBONATE ION, Membrane-associated protein SbtB, SODIUM ION, ... | | Authors: | Fang, S, Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8FG7
 
 | |
7E1H
 
 | | crystal structure of RD-BEF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA-binding response regulator, MAGNESIUM ION | | Authors: | Hong, S, Zhang, X, Zhang, P. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
7TCZ
 
 | | Human cytomegalovirus protease mutant (C84A, C87A, C138A, C202A) in complex with inhibitor | | Descriptor: | Assemblin, [1-(2-oxopropyl)-4-phenyl-1H-1,2,3-triazol-5-yl]methyl benzylcarbamate | | Authors: | Hulce, K.R, Bohn, M, Ongpipattanakul, C, Jaishankar, P, Renslo, A.R, Craik, C.S. | | Deposit date: | 2021-12-29 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Inhibiting a dynamic viral protease by targeting a non-catalytic cysteine.
Cell Chem Biol, 29, 2022
|
|
8FOR
 
 | |
8FMC
 
 | |
8FMD
 
 | |
8FOQ
 
 | |
8FME
 
 | |
8FOS
 
 | |
6WY3
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; P212121 form | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*(LV2)P*GP*CP*CP*GP*G)-3') | | Authors: | Sekula, B, Ruszkowski, M, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6WY2
 
 | | Crystal structure of RNA-10mer: CCGG(N4-methyl-C)GCCGG | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA-10mer: CCGG(4-methyl-C)GCCGG | | Authors: | Sekula, B, Ruszkowski, M, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
4OV9
 
 | | Structure of isopropylmalate synthase binding with alpha-isopropylmalate | | Descriptor: | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid, ZINC ION, isopropylmalate synthase | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
5XSD
 
 | | XylFII-LytSN complex mutant - D103A | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8Q71
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
7CX4
 
 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
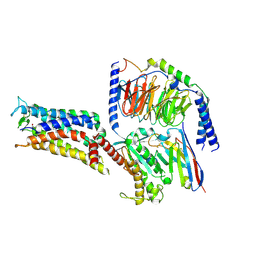 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
4O1I
 
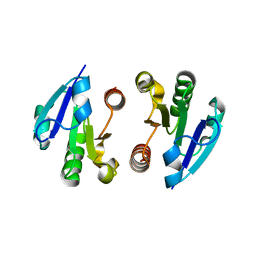 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
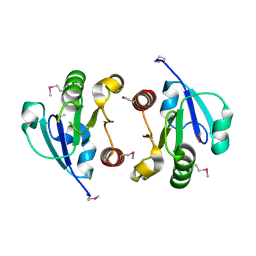 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4I0C
 
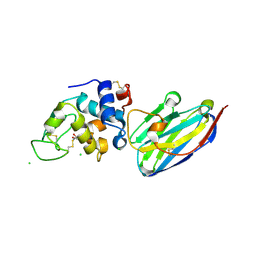 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
