1LW7
 
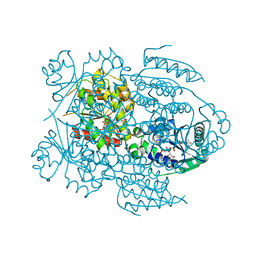 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
3G5A
 
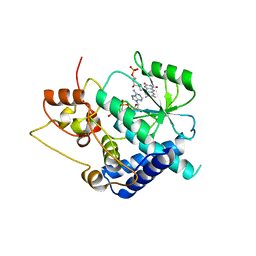 | |
4I5X
 
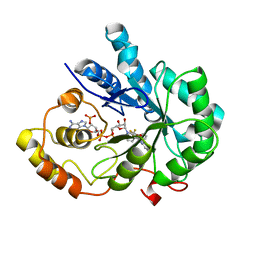 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
3WVJ
 
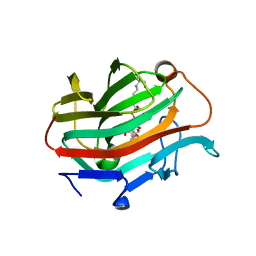 | | The crystal structure of native glycosidic hydrolase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | Authors: | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
3WRF
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
1Q9S
 
 | | Crystal structure of riboflavin kinase with ternary product complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-08-25 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism.
Biochemistry, 42, 2003
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
4E5K
 
 | |
7CQZ
 
 | | crystal structure of mouse FAM46C (TENT5C) | | Descriptor: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | Authors: | Hu, J.L, Zhang, H, Gao, S. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34969759 Å) | | Cite: | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
8THO
 
 | |
1M4M
 
 | | Mouse Survivin | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, ZINC ION | | Authors: | Muchmore, S.W, Chen, J, Jakob, C, Zakula, D, Matayoshi, E.D, Wu, W, Zhang, H, Li, F, Ng, S.C, Altieri, D.C. | | Deposit date: | 2002-07-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE AND MUTAGENIC ANALYSIS OF THE INHIBITOR-OF-APOPTOSIS PROTEIN SURVIVIN
MOL.CELL, 6, 2000
|
|
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
8GZC
 
 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
3GZ5
 
 | | Crystal structure of Shewanella oneidensis NrtR | | Descriptor: | MutT/nudix family protein, SODIUM ION | | Authors: | Huang, N, Zhang, H. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism
Structure, 17, 2009
|
|
7V6D
 
 | | Structure of lipase B from Lasiodiplodia theobromae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lipase B | | Authors: | Xue, B, Zhang, H.F, Nguyen, G.K.T, Yew, W.S. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides' Precursors.
Int J Mol Sci, 22, 2021
|
|
1J85
 
 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
5YEP
 
 | | Crystal structure of SO_3166-SO_3165 from Shewanella oneidensis | | Descriptor: | Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Jia, X, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-28 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system.
J. Biol. Chem., 293, 2018
|
|
7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E32
 
 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2X
 
 | | Apo serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7D77
 
 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7JLV
 
 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Disease resistance protein Roq1, MAGNESIUM ION | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
5ZYL
 
 | | Crystal structure of CERT START domain in complex with compound E25A | | Descriptor: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
