7MDS
 
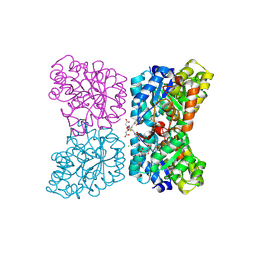 | | Crystal structure of AtDHDPS1 in complex with MBDTA-2 | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase 1, chloroplastic, CHLORIDE ION, ... | | Authors: | Hall, C.J, Soares da Costa, T.P, Panjikar, S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants.
Elife, 10, 2021
|
|
3TCM
 
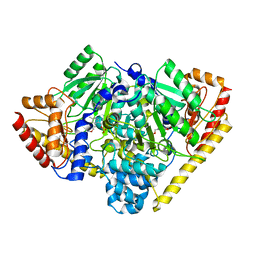 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
6OVB
 
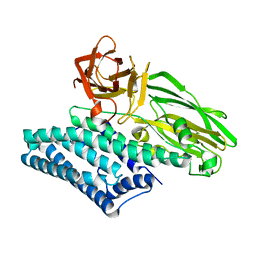 | | Crystal structure of a Bacillus thuringiensis Cry1Da tryptic core variant | | Descriptor: | Active core crystal toxin protein 1D | | Authors: | Rydel, T.J, Halls, C, Evdokimov, A.G, Beishir, S.C. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Bacillus thuringiensis Cry1Da_7 and Cry1B.868 Protein Interactions with Novel Receptors Allow Control of Resistant Fall Armyworms, Spodoptera frugiperda (J.E. Smith).
Appl.Environ.Microbiol., 85, 2019
|
|
6D3J
 
 | | FT_T dioxygenase holoenzyme | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FT_T dioxygenase | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6D3M
 
 | | FT_T dioxygenase with bound quizalofop | | Descriptor: | (2R)-2-{4-[(6-chloroquinoxalin-2-yl)oxy]phenoxy}propanoic acid, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6D3I
 
 | | ftv7 dioxygenase with 2,4-D bound | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6D3H
 
 | | FT_T dioxygenase with bound dichlorprop | | Descriptor: | (2R)-2-(2,4-dichlorophenoxy)propanoic acid, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6D1O
 
 | | FT_5 dioxygenase apoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
6VVI
 
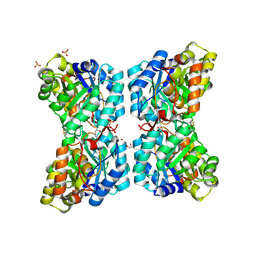 | |
6VVH
 
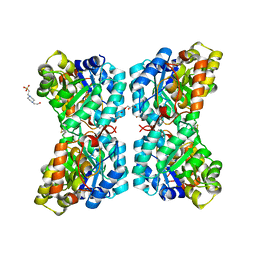 | |
6P90
 
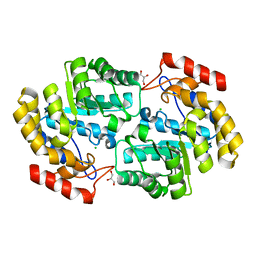 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
8EZ0
 
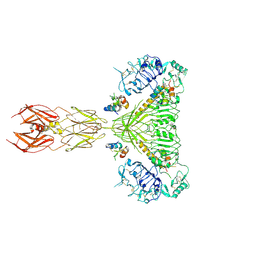 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Symmetric conformation | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYX
 
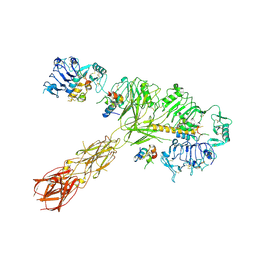 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S; denoted as IR-3CS) Asymmetric conformation 1 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYY
 
 | | Cryo-EM structure of 4 insulins bound full-length mouse IR mutant with physically decoupled alpha CTs (C684S/C685S/C687S, denoted as IR-3CS) Asymmetric conformation 2 | | Descriptor: | Insulin, Insulin receptor | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8EYR
 
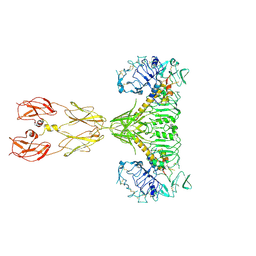 | | Cryo-EM structure of two IGF1 bound full-length mouse IGF1R mutant (four glycine residues inserted in the alpha-CT; IGF1R-P674G4): symmetric conformation | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Wu, J.Y, Hall, C, Bai, X.C, Choi, E. | | Deposit date: | 2022-10-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis for the role of disulfide-linked alpha CTs in the activation of insulin-like growth factor 1 receptor and insulin receptor.
Elife, 11, 2022
|
|
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
7TYJ
 
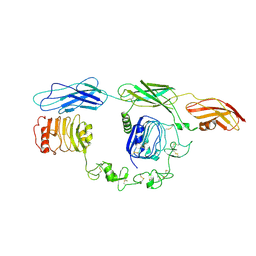 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
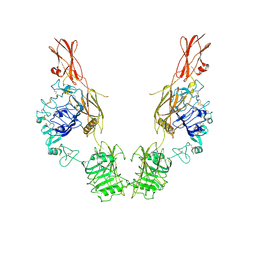 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
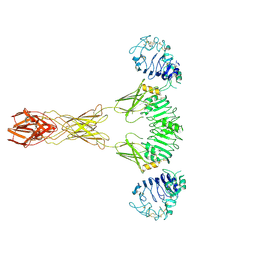 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | Descriptor: | Insulin receptor-related protein | | Authors: | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2KH2
 
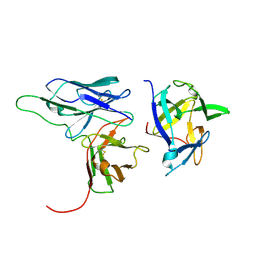 | | Solution structure of a scFv-IL-1B complex | | Descriptor: | Interleukin-1 beta, scFv | | Authors: | Wilkinson, I.C, Hall, C.J, Veverka, V, Muskett, F.W, Stephens, P.E, Taylor, R.J, Henry, A.J, Carr, M.D. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | High resolution NMR-based model for the structure of a scFv-IL-1beta complex: potential for NMR as a key tool in therapeutic antibody design and development.
J.Biol.Chem., 284, 2009
|
|
8DTM
 
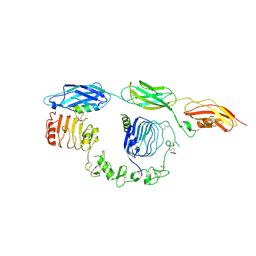 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | Descriptor: | Insulin mimetic peptide S597 component 2, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DTL
 
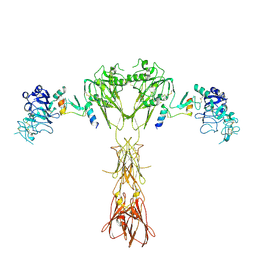 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8U4B
 
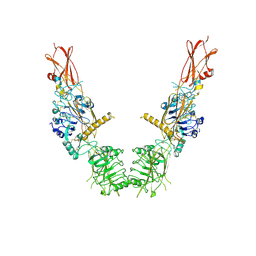 | | Cryo-EM structure of long form insulin receptor (IR-B) in the apo state | | Descriptor: | Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJB
 
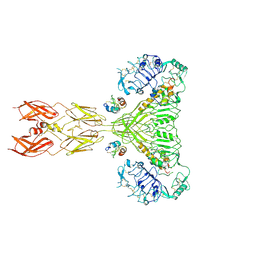 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8VJC
 
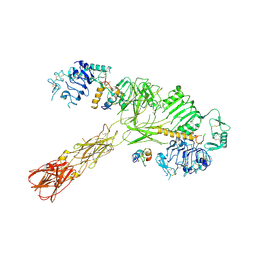 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
