8VTR
 
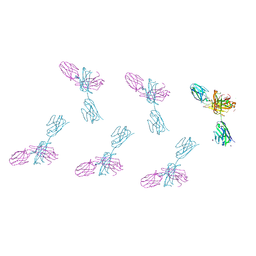 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VU4
 
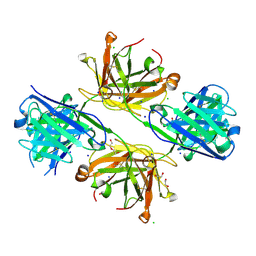 | | Structure of FabS1CE4-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VU1
 
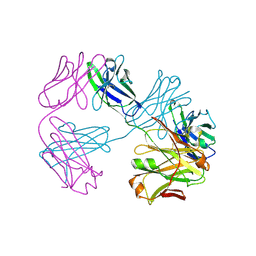 | | Structure of FabS1CE3-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor (trigonal form) | | Descriptor: | S1CE3 VARIANT OF FAB-EPR-1 heavy chain, S1CE3 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VTP
 
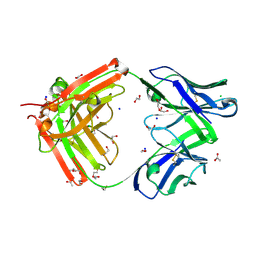 | | Structure of FabS1CE-EPR-1, a high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VVO
 
 | | Structure of FabS1CE2-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | CHLORIDE ION, Erythropoietin receptor, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUI
 
 | | Structure of FabS1CE-EPR-1, an elbow-locked Fab, in complex with the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
8VUC
 
 | | Structure of FabS1CE2-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE2 VARIANT OF FAB-EPR-1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8VUA
 
 | | Structure of FabS1CE1-EPR-1, an elbow-locked high affinity antibody for the erythropoeitin receptor | | Descriptor: | S1CE1 VARIANT OF FAB-EPR-1 heavy chain, S1CE1 VARIANT OF FAB-EPR-1 light chain | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
8SWL
 
 | |
8CZV
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZU
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 16d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{S})-2-(cyclohexylmethyl)cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZT
 
 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZX
 
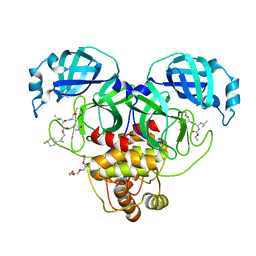 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CZW
 
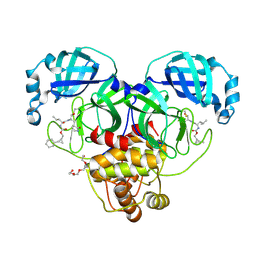 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
1U3R
 
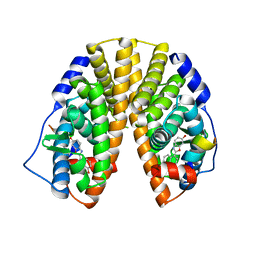 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-338 | | Descriptor: | 2-(5-HYDROXY-NAPHTHALEN-1-YL)-1,3-BENZOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
1U9E
 
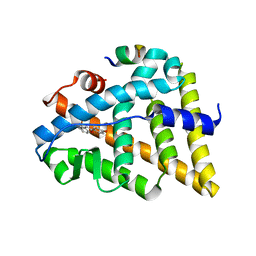 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-397 | | Descriptor: | 2-(4-HYDROXY-PHENYL)BENZOFURAN-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design Of Estrogen Receptor-beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1U3S
 
 | | Crystal Structure of Estrogen Receptor beta complexed with WAY-797 | | Descriptor: | 3-(6-HYDROXY-NAPHTHALEN-2-YL)-BENZO[D]ISOOXAZOL-6-OL, Estrogen receptor beta, steroid receptor coactivator-1 | | Authors: | Malamas, M.S, Manas, E.S, McDevitt, R.E, Gunawan, I, Xu, Z.B, Collini, M.D, Miller, C.P, Dinh, T, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of aryl diphenolic azoles as potent and selective estrogen receptor-beta ligands.
J.Med.Chem., 47, 2004
|
|
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7I
 
 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
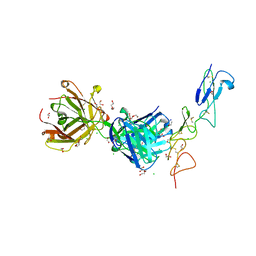 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
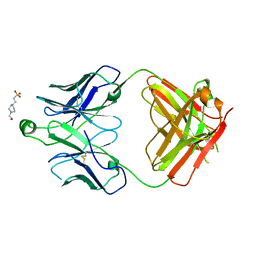 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
2IL6
 
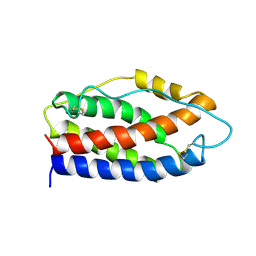 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
