5UEN
 
 | | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution | | Descriptor: | 4-{[3-(8-cyclohexyl-2,6-dioxo-1-propyl-1,2,6,7-tetrahydro-3H-purin-3-yl)propyl]carbamoyl}benzene-1-sulfonyl fluoride, Adenosine receptor A1,Soluble cytochrome b562,Adenosine receptor A1, OLEIC ACID | | Authors: | Glukhova, A, Thal, D.M, Nguyen, A.T, Vecchio, E.A, Jorg, M, Scammells, P.J, May, L.T, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity.
Cell, 168, 2017
|
|
4X96
 
 | |
4X97
 
 | |
4X95
 
 | |
4X92
 
 | |
4X93
 
 | | Crystal structure of Lysosomal Phospholipase A2 crystallized in the presence of methyl arachidonyl fluorophosphonate (tetragonal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
4X94
 
 | |
4X91
 
 | |
4X90
 
 | | Crystal structure of Lysosomal Phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase.
Nat Commun, 6, 2015
|
|
6E3Y
 
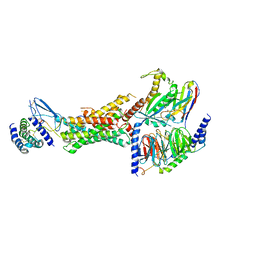 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
7RR0
 
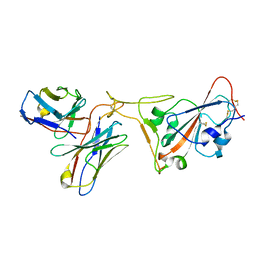 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | Authors: | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | Deposit date: | 2021-08-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
6NIY
 
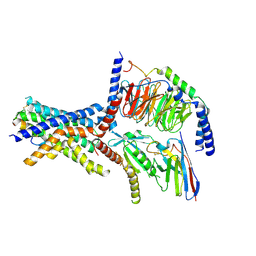 | | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | dal Maso, E, Glukhova, A, Zhu, Y, Garcia-Nafria, J, Tate, C.G, Atanasio, S, Reynolds, C.A, Ramirez-Aportela, E, Carazo, J.-M, Hick, C.A, Furness, S.G.B, Hay, D.L, Liang, Y.-L, Miller, L.J, Christopoulos, A, Wang, M.-W, Wootten, D, Sexton, P.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-01-23 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | The Molecular Control of Calcitonin Receptor Signaling.
Acs Pharmacol Transl Sci, 2, 2019
|
|
6D9H
 
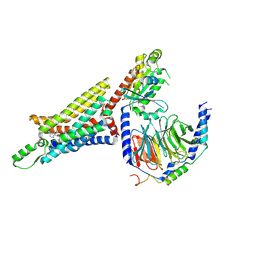 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Khoshouei, M, Thal, D.M, Liang, Y.-L, Nguyen, A.T.N, Furness, S.G.B, Venugopal, H, Baltos, J, Plitzko, J.M, Danev, R, Baumeister, W, May, L.T, Wootten, D, Sexton, P, Glukhova, A, Christopoulos, A. | | Deposit date: | 2018-04-29 | | Release date: | 2018-06-20 | | Last modified: | 2018-07-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the adenosine-bound human adenosine A1receptor-Gicomplex.
Nature, 558, 2018
|
|
8GHD
 
 | | The structure of h12-LOX in hexameric form bound to inhibitor ML355 and arachidonic acid | | Descriptor: | ARACHIDONIC ACID, FE (II) ION, N-(1,3-benzothiazol-2-yl)-4-{[(2-hydroxy-3-methoxyphenyl)methyl]amino}benzene-1-sulfonamide, ... | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHC
 
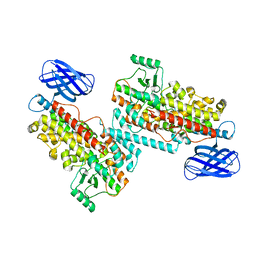 | | The structure of h12-LOX in dimeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHE
 
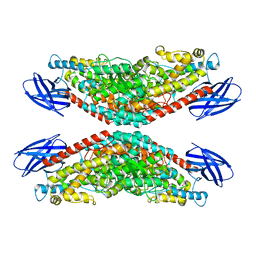 | | The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
8GHB
 
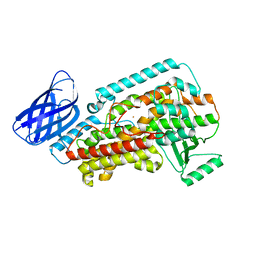 | | The structure of h12-LOX in monomeric form | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12 | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
8FX5
 
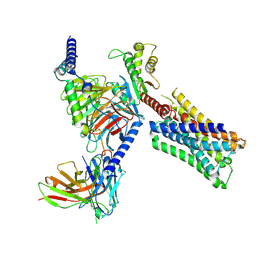 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and xanomeline | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2023-01-23 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Xanomeline displays concomitant orthosteric and allosteric binding modes at the M 4 mAChR.
Nat Commun, 14, 2023
|
|
5TXF
 
 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in a closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phosphatidylcholine-sterol acyltransferase, ... | | Authors: | Manthei, K.A, Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A retractable lid in lecithin:cholesterol acyltransferase provides a structural mechanism for activation by apolipoprotein A-I.
J. Biol. Chem., 292, 2017
|
|
7LD3
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7LD4
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7LX5
 
 | |
7T3X
 
 | | Structure of unphosphorylated Pediculus humanus (Ph) PINK1 D334A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1 | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
