5OCN
 
 | | Crystal structure of the forkhead domain of human FOXN1 | | Descriptor: | Forkhead box protein N1, POTASSIUM ION | | Authors: | Newman, J.A, Aitkenhead, H, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-07-03 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the forkhead domain of human FOXN1
To be published
|
|
5NZY
 
 | | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, DNA cross-link repair 1A protein, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Kupinska, K, Burgess-Brown, N.A, Talon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime
To Be Published
|
|
4WSQ
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
5LB3
 
 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, MAGNESIUM ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
5LB8
 
 | | Crystal structure of human RECQL5 helicase APO form. | | Descriptor: | ATP-dependent DNA helicase Q5, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
2P0A
 
 | | The crystal structure of human synapsin III (SYN3) in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Turnbull, A.P, Phillips, C, Pike, A.C.W, Elkins, J.M, Gileadi, C, Salah, E, Niesen, F.H, Burgess, N, Gileadi, O, Gorrec, F, Umeano, C, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human synapsin III (SYN3) in complex with AMPPNP
To be Published
|
|
6ZSL
 
 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
6HQ9
 
 | | Crystal structure of the Tudor domain of human ERCC6-L2 | | Descriptor: | DNA excision repair protein ERCC-6-like 2 | | Authors: | Newman, J.A, Gavard, A.E, Nathan, W.J, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-09-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structure of the Tudor domain of human ERCC6-L2
To Be Published
|
|
6ZU8
 
 | | Crystal structure of human Brachyury G177D variant in complex with Afatinib | | Descriptor: | Brachyury protein, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human Brachyury G177D variant in complex with Afatinib
To Be Published
|
|
6IBE
 
 | | The FERM domain of Human EPB41L3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Band 4.1-like protein 3 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The FERM domain of Human EPB41L3
To Be Published
|
|
6IBD
 
 | | The Phosphatase and C2 domains of Human SHIP1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
7ABS
 
 | | Structure of human DCLRE1C/Artemis in complex with DNA - re-evaluation of 6WO0 | | Descriptor: | DNA (5'-D(*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*GP*CP*T)-3'), Protein artemis, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6CNH
 
 | | Human PRPF4B in complex with Rebastinib | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Godoi, P.H.C, Santiago, A.S, Ramos, P.Z, Fala, A.M, Salmazo, A.P.T, Counago, R.M, Righetto, G.L, Silva, P.N.B, Gileadi, O, Guimaraes, C.R.W, Massirer, K.B, Arruda, P, Elkins, J.M, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human PRPF4B in complex with Rebastinib
To be Published
|
|
7AFS
 
 | | The structure of Artemis variant D37A | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AGI
 
 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7B2X
 
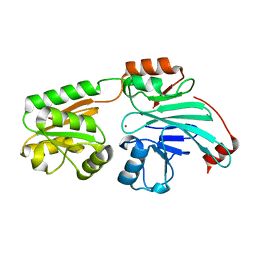 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7AW6
 
 | | The extracellular region of CD33 with bound sialoside analogue P22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Thompson, A.P, Arrowsmith, C.H, Edwards, A.M, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The extracellular region of CD33 with bound sialoside analogue P22
To Be Published
|
|
7APV
 
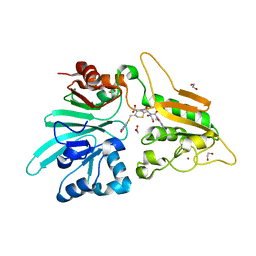 | | Structure of Artemis/DCLRE1C/SNM1C in complex with Ceftriaxone | | Descriptor: | 1,2-ETHANEDIOL, Ceftriaxone, NICKEL (II) ION, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AFU
 
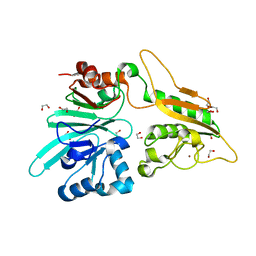 | | The structure of Artemis variant H33A | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
5UV4
 
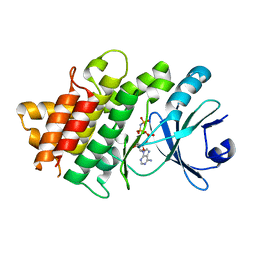 | | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Putative leucine-rich repeat protein kinase family protein | | Authors: | Counago, R.M, Aquino, B, Massirer, K.B, Gileadi, O, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP
To Be Published
|
|
7AF1
 
 | | The structure of Artemis/SNM1C/DCLRE1C with 2 Zinc ions | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
2REY
 
 | | Crystal structure of the PDZ domain of human dishevelled 2 (homologous to Drosophila dsh) | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Papagrigoriou, E, Gileadi, C, Elkins, J, Cooper, C, Ugochukwu, E, Turnbull, A, Pike, A.C.W, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the PDZ domains of human dishevelled 2 (homologous to Drosophila dsh).
To be Published
|
|
5LBA
 
 | | Crystal structure of human RECQL5 helicase in complex with DSPL fragment(1-cyclohexyl-3-(oxolan-2-ylmethyl)urea, SGC - Diamond XChem I04-1 fragment screening. | | Descriptor: | 1-cyclohexyl-3-[[(2~{R})-oxolan-2-yl]methyl]urea, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, ... | | Authors: | Newman, J.A, Aitkenhead, H, Talon, R, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human RECQL5 helicase in complex with 3D fragment (1-cyclohexyl-3-(oxolan-2-ylmethyl)urea)
To be published
|
|
5LB5
 
 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg (tricilinc form). | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, DIMETHYL SULFOXIDE, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
7A1F
 
 | | Crystal structure of human 5' exonuclease Appollo in complex with 5'dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5' exonuclease Apollo, FE (III) ION, ... | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
