6N5K
 
 | | Structure of Human pir-miRNA-449c Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
6N5O
 
 | | Structure of Human pir-miRNA-202 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
5JDV
 
 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JEH
 
 | | Human carbonic anhydrase II (L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JG3
 
 | | Human carbonic anhydrase II (121T/N67Q) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JGS
 
 | | Human carbonic anhydrase II (F131Y/L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6N5T
 
 | | Structure of Human pir-miRNA-378a Apical Loop Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
5KMQ
 
 | |
5L03
 
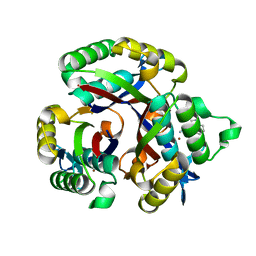 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | Authors: | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
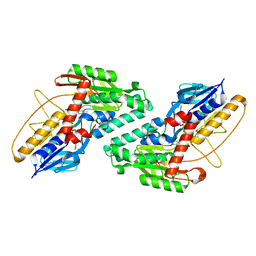
5KMP
 
 | |
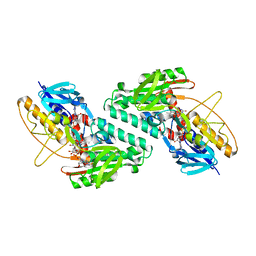
5KMR
 
 | |
6NZ9
 
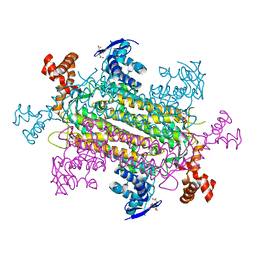 | | Crystal structure of E. coli fumarase C bound to citrate at 1.53 angstrom resolution | | Descriptor: | CITRIC ACID, Fumarate hydratase class II | | Authors: | Stuttgen, G.M, May, J.F, Bhattcharyya, B, Weaver, T.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Closed fumarase C active-site structures reveal SS Loop residue contribution in catalysis.
Febs Lett., 594, 2020
|
|
3IF6
 
 | | Crystal structure of OXA-46 beta-lactamase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, L(+)-TARTARIC ACID, ... | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Giuliani, F, Kapetis, D, De Luca, F, Rossolini, G.M, Mangani, S. | | Deposit date: | 2009-07-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the narrow-spectrum OXA-46 class D beta-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates
Antimicrob.Agents Chemother., 54, 2010
|
|
7XMV
 
 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A(AMP/ADP) filament bound with ADP, AMP and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XN3
 
 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type B filament bound with Pi | | Descriptor: | PHOSPHATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XMU
 
 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filament bound with ADP, Pi and R5P | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7UJ4
 
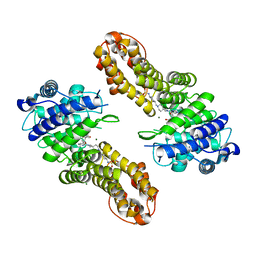 | | Inhibition of Human Menin by SNDX-5613 | | Descriptor: | 2-({4-[7-({(1r,4r)-4-[(ethanesulfonyl)amino]cyclohexyl}methyl)-2,7-diazaspiro[3.5]nonan-2-yl]pyrimidin-5-yl}oxy)-N-ethyl-5-fluoro-N-(propan-2-yl)benzamide, Isoform 2 of Menin, MAGNESIUM ION | | Authors: | McKeever, B.M, Kulkarni, S, McGeehan, G.M. | | Deposit date: | 2022-03-30 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | MEN1 mutations mediate clinical resistance to menin inhibition.
Nature, 615, 2023
|
|
7WV3
 
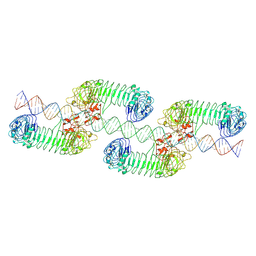 | | Toll-like receptor3 linear cluster | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (80-MER), ... | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVF
 
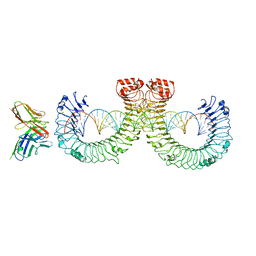 | | ectoTLR3-mAb12-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3, mAb12 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVJ
 
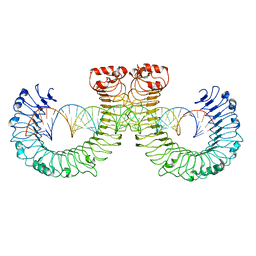 | | NT-mut(K117D,K139D,K145D) TLR3 -poly I:C complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV5
 
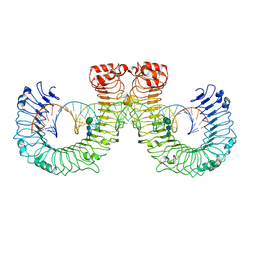 | | ectoTLR3-poly(I:C) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (46-MER), ... | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV4
 
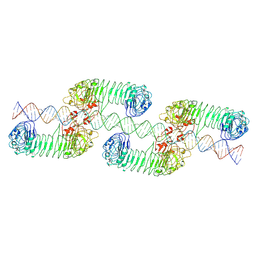 | | ectoTLR3-poly(I:C) cluster | | Descriptor: | RNA (80-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVE
 
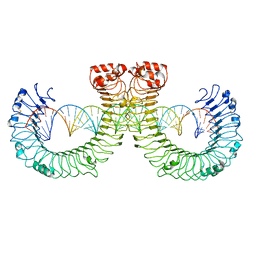 | | CT-mut (D523K,D524K,E527K) TLR3-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7UDT
 
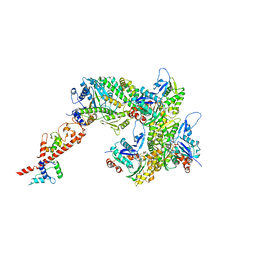 | | cryo-EM structure of the rigor state wild type myosin-15-F-actin complex (symmetry expansion and re-centering) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
7UDU
 
 | | cryo-EM structure of the ADP state wild type myosin-15-F-actin complex (symmetry expansion and re-centering) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gong, R, Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-03-20 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia.
Sci Adv, 8, 2022
|
|
