3MBC
 
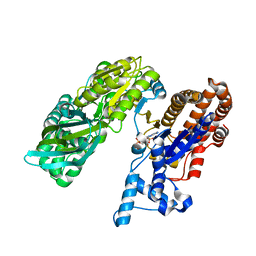 | | Crystal structure of monomeric isocitrate dehydrogenase from Corynebacterium glutamicum in complex with NADP | | Descriptor: | Isocitrate dehydrogenase [NADP], MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sidhu, N.S, Aich, S, Sheldrick, G.M, Delbaere, L.T.J. | | Deposit date: | 2010-03-25 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly NADP+-specific isocitrate dehydrogenase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7AF2
 
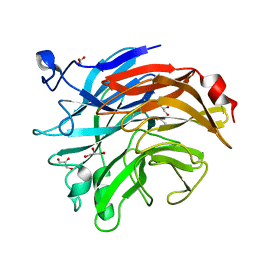 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
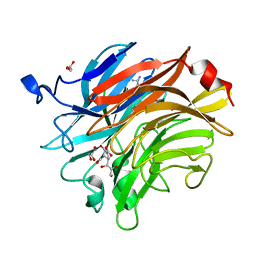 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
5L87
 
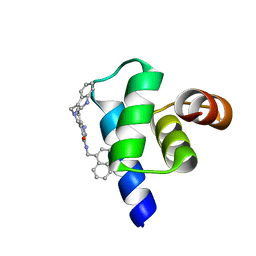 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-indol-3-ylmethyl)-1-methyl-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, Peroxin 14 | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
5KMS
 
 | |
5L8A
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(phenylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, GLYCINE, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
5EEQ
 
 | | Grb7 SH2 with the G7-B1 bicyclic peptide inhibitor | | Descriptor: | Bicyclic Peptide Inhibitor, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Ambaye, N.D, Watson, G.M, Wilce, M.C.J, Wilce, G.M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
6DQX
 
 | | Actinobacillus ureae class Id ribonucleotide reductase alpha subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McBride, M.J, Palowitch, G.M, Boal, A.K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis.
Biochemistry, 58, 2019
|
|
6D8L
 
 | |
6DKU
 
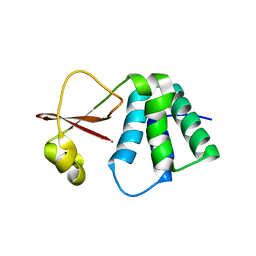 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
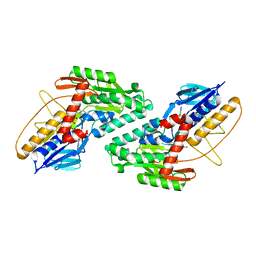
5WED
 
 | |
6D8N
 
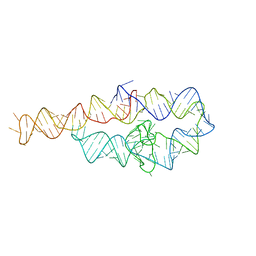 | |
2YHH
 
 | | Microvirin:mannobiose complex | | Descriptor: | MANNAN-BINDING LECTIN, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hussan, S, Bewley, C.A, Clore, G.M, Gustchina, E, Ghirlando, R. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Monovalent Lectin Microvirin in Complex with Man(Alpha)(1-2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity.
J.Biol.Chem., 286, 2011
|
|
2YBA
 
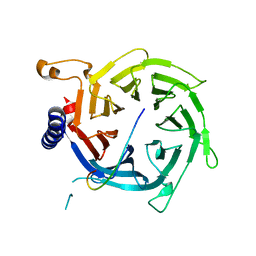 | | Crystal structure of Nurf55 in complex with histone H3 | | Descriptor: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
2YKA
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HVS ORF57 peptide | | Descriptor: | 52 KDA IMMEDIATE-EARLY PHOSPHOPROTEIN, RNA AND EXPORT FACTOR-BINDING PROTEIN 2 | | Authors: | Tunnicliffe, R.B, Hautbergue, G.M, Kalra, P, Wilson, S.A, Golovanov, A.P. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri Orf57 to the Mammalian Export Adaptor Alyref.
Plos Pathog., 10, 2014
|
|
2YAJ
 
 | | CRYSTAL STRUCTURE OF GLYCYL RADICAL ENZYME with bound substrate | | Descriptor: | 4-HYDROXYPHENYLACETATE, 4-HYDROXYPHENYLACETATE DECARBOXYLASE LARGE SUBUNIT, 4-HYDROXYPHENYLACETATE DECARBOXYLASE SMALL SUBUNIT, ... | | Authors: | Martins, B.M, Blaser, M, Feliks, M, Ullmann, G.M, Selmer, T. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Structural Basis for a Kolbe-Type Decarboxylation Catalyzed by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 133, 2011
|
|
4ZNF
 
 | |
2YPK
 
 | | Structural features underlying T-cell receptor sensitivity to concealed MHC class I micropolymorphisms | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-57 ALPHA CHAIN, ... | | Authors: | Stewart-Jones, G.B, Simpson, P, Van Der Merwe, P.A, Easterbrook, P, Mcmichael, A.J, Rowland-Jones, S.L, Jones, E.Y, Gillespie, G.M. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Features Underlying T-Cell Receptor Sensitivity to Concealed Mhc Class I Micropolymorphisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
2B8J
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with adenosine and phosphate at 2 A resolution | | Descriptor: | ADENOSINE, GOLD 3+ ION, GOLD ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Thaller, M.C, Rossolini, G.M, Mangani, S. | | Deposit date: | 2005-10-07 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | A structure-based proposal for the catalytic mechanism of the bacterial acid phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases
J.Mol.Biol., 355, 2006
|
|
5ADP
 
 | | Crystal structure of the A.17 antibody FAB fragment - Light chain S35R mutant | | Descriptor: | FAB A.17 | | Authors: | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | Deposit date: | 2015-08-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
8JA0
 
 | |
8Q34
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the ligand ZZ001229a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1~{H}-imidazo[4,5-b]pyridin-2-ylmethyl)-3-(3-methyl-1,2-diazirin-3-yl)propanamide | | Authors: | MacLean, E.M, Gao, Q, Williams, E, Balcomb, B.H, von Delft, F, Bajusz, D, Keeley, A, Abranyi-Balogh, P, Koekemoer, L, Keseru, G.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein binding sites by photoreactive fragment pharmacophores.
Commun Chem, 7, 2024
|
|
1RMT
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase complexed with adenosine. | | Descriptor: | ADENOSINE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
7MMT
 
 | |
