6G0Y
 
 | | X-ray structure of M-21 protein complex | | Descriptor: | Matrix M2-1, Phosphoprotein, ZINC ION | | Authors: | Edwards, T.A, Barr, J. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structure of the Human Respiratory Syncytial Virus M2-1 Protein Bound to the Interaction Domain of the Phosphoprotein P Defines the Orientation of the Complex.
Mbio, 9, 2018
|
|
3H3D
 
 | |
1Q7F
 
 | |
2FE9
 
 | |
7QGS
 
 | | Crystal structure of p300 CH1 domain in complex with CITIF (a CITED2-HIF-1alpha hybrid) | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone acetyltransferase, ZINC ION | | Authors: | Hegedus, Z, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding p300-transcription factor interactions using sequence variation and hybridization.
Rsc Chem Biol, 3, 2022
|
|
7NY8
 
 | | Affimer K69 - KRAS protein complex | | Descriptor: | Affimer K69, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turner, A.L, Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2021-03-21 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
8CBW
 
 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly monomer | | Descriptor: | Nucleocapsid, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZLA
 
 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
5IU1
 
 | | N-terminal PAS domain homodimer of PpANR MAP3K from Physcomitrella patens. | | Descriptor: | CTR1-like protein, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stevenson, R.R, Trinh, C.H, Edwards, T.A, Cuming, A.C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance.
Plant Cell, 28, 2016
|
|
4AKL
 
 | | Structure of the Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein | | Descriptor: | NUCLEOCAPSID, TRIETHYLENE GLYCOL | | Authors: | Carter, S.D, Walter, C.T, Surtees, R, Bergeron, E, Ariza, A, Albarino, C.G, Nichol, S.T, Hiscox, J.A, Edwards, T.A, Barr, J.N. | | Deposit date: | 2012-02-24 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, Function, and Evolution of the Crimean-Congo Hemorrhagic Fever Virus Nucleocapsid Protein.
J.Virol., 86, 2012
|
|
8C4H
 
 | | CryoEM structure of the Hendra henipavirus nucleocapsid sauronoid assembly multimer | | Descriptor: | Nucleocapsid, RNA (84-MER) | | Authors: | Passchier, T.C, Maskell, D.P, Edwards, T.A, Barr, J.N. | | Deposit date: | 2023-01-04 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.485 Å) | | Cite: | The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Sci Rep, 14, 2024
|
|
6HJL
 
 | | Affimer:BclxL | | Descriptor: | Affimer ADB13, Bcl-2-like protein 1 | | Authors: | Miles, J.A, Trinh, C.H, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2018-09-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs
Biorxiv, 2020
|
|
6YR8
 
 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
6YXW
 
 | | Affimer K3 - KRAS protein complex | | Descriptor: | Affimer K3, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
5ELJ
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 1 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5EQL
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5ELU
 
 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SULFATE ION, SUMO-Affirmer-S2B3, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-05 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
5A0O
 
 | | adhiron raised against p300 | | Descriptor: | ADHIRON | | Authors: | Kyle, H.F, Wickson, K.F, Stott, J, Burslem, G.M, Breeze, A.L, Tiede, C, Tomlinson, D.C, Warriner, S.L, Nelson, A, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Exploration of the Hif-1Alpha.P300 Interface Using Peptide and Adhiron Phage Display Technologies
Mol.Biosyst., 11, 2015
|
|
5C3F
 
 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
5A37
 
 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | HUMAN ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
5A36
 
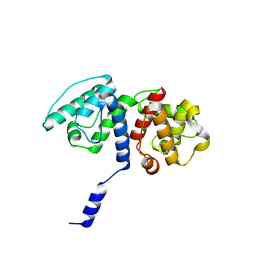 | | Mutations in the Calponin homology domain of Alpha-Actinin-2 affect Actin binding and incorporation in muscle. | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Haywood, N.J, Wolny, M, Trinh, C.H, Shuping, Y, Edwards, T.A, Peckham, M. | | Deposit date: | 2015-05-27 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypertrophic Cardiomyopathy Mutations in the Calponin-Homology Domain of Actn2 Affect Actin Binding and Cardiomyocyte Z-Disc Incorporation.
Biochem.J., 473, 2016
|
|
6ST2
 
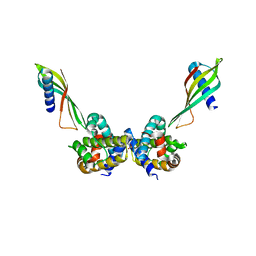 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Affimer AF6, Bcl-2-like protein 1, SULFATE ION | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
5A97
 
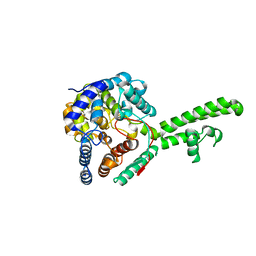 | | Hazara virus nucleocapsid protain | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Surtees, R, Ariza, A, Hewson, R, Barr, J.N, Edwards, T.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Hazara Virus Nucleocapsid Protein.
Bmc Struct.Biol., 15, 2015
|
|
