4HQU
 
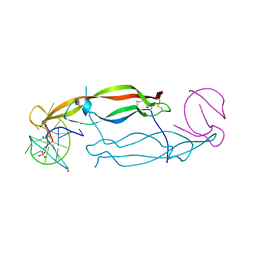 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KMS
 
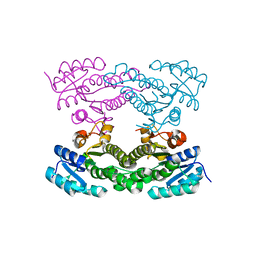 | | Crystal structure of Acetoacetyl-CoA reductase from Rickettsia felis | | Descriptor: | Acetoacetyl-CoA reductase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J, Lukacs, C, Edwards, T.E, Lorimer, D. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetoacetyl-CoA reductase from Rickettsia felis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6MWE
 
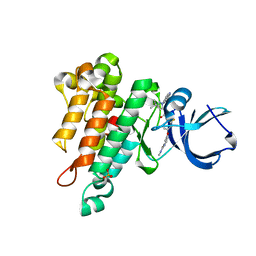 | | CRYSTAL STRUCTURE OF TIE2 IN COMPLEX WITH DECIPERA COMPOUND DP1919 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Angiopoietin-1 receptor, SULFATE ION | | Authors: | Chun, L, Abendroth, J, Edwards, T.E. | | Deposit date: | 2018-10-29 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Selective Tie2 Inhibitor Rebastinib Blocks Recruitment and Function of Tie2
Mol. Cancer Ther., 16, 2017
|
|
4WMR
 
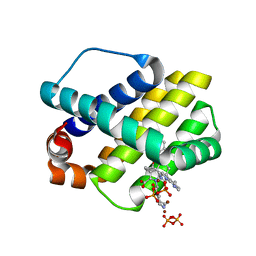 | | STRUCTURE OF MCL1 BOUND TO BRD inhibitor ligand 1 AT 1.7A | | Descriptor: | 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, PYROPHOSPHATE 2-, ... | | Authors: | CLIFTON, M.C, EDWARDS, T.E. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6VC8
 
 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
6OOZ
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (S)-{1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, CHLORIDE ION, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fox III, D, Lecomte, F, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
6OP0
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (R)-{1-[(2,5-dimethylphenyl)methyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-benzimidazol-2-yl}(pyridin-4-yl)methanol, Tumor necrosis factor | | Authors: | Arakaki, T.L, Edwards, T.E, Fairman, J.W, Davies, D.R, Foley, A, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
3K1V
 
 | |
6OOY
 
 | | Asymmetric hTNF-alpha | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Tumor necrosis factor, {1-[(2,5-dimethylphenyl)methyl]-1H-benzimidazol-2-yl}methanol | | Authors: | Arakaki, T.L, Edwards, T.E, Ceska, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Small molecules that inhibit TNF signalling by stabilising an asymmetric form of the trimer.
Nat Commun, 10, 2019
|
|
3K0J
 
 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
3EGZ
 
 | | Crystal structure of an in vitro evolved tetracycline aptamer and artificial riboswitch | | Descriptor: | 7-CHLOROTETRACYCLINE, MAGNESIUM ION, Tetracycline aptamer and artificial riboswitch, ... | | Authors: | Xiao, H, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch
Chem.Biol., 15, 2008
|
|
7TI2
 
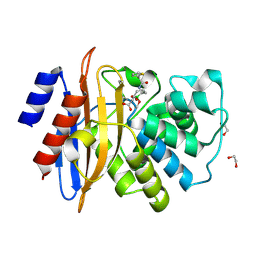 | | Structure of KPC-2 bound to RPX-7063 at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolyzing beta-lactamase KPC, {(3R,7S)-2-hydroxy-3-[2-(thiophen-2-yl)acetamido]-2,3,4,7-tetrahydro-1,2-oxaborepin-7-yl}acetic acid | | Authors: | Clifton, M.C, Fairman, J.W, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
7TI1
 
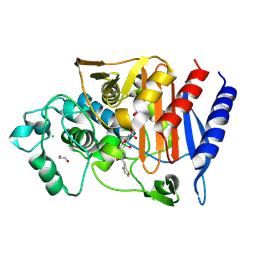 | | Structure of AmpC bound to RPX-7063 at 2.0A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
7TI0
 
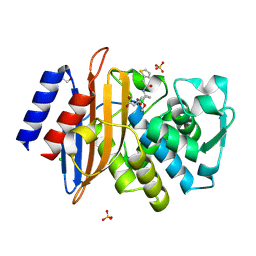 | | Structure of CTX-M-15 bound to RPX-7063 at 1.5A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Abendroth, J, Hecker, S.J, Edwards, T.E. | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
3STH
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Toxoplasma gondii | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Edwards, T.E, Sankaran, B. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Membrane skeletal association and post-translational allosteric regulation of Toxoplasma gondii GAPDH1.
Mol.Microbiol., 103, 2017
|
|
4NW7
 
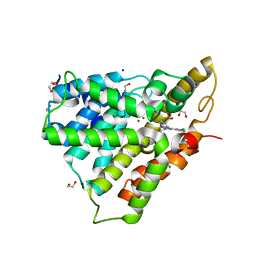 | | PDE4 catalytic domain | | Descriptor: | (4-{[4-(3-chlorophenyl)-6-cyclopropyl-1,3,5-triazin-2-yl]amino}phenyl)acetic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Fox III, D, Edwards, T.E. | | Deposit date: | 2013-12-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of triazines as selective PDE4B versus PDE4D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4MYQ
 
 | |
5VRM
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3-oxidanyl-4-[[1-oxidanyl-6-[4-(trifluoromethyl)phenoxy]-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5VRL
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5VRN
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-4-[[5-[4-cyano-2-[(~{E})-hydroxyiminomethyl]phenoxy]-1-oxidanyl-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
5W07
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-30 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor of Mycobacterium tuberculosis InhA
To Be Published
|
|
6CKG
 
 | | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487) | | Descriptor: | 1,2-ETHANEDIOL, D-glycerate 3-kinase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487)
To be Published
|
|
6CAU
 
 | | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-31 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | UDP-N-acetylmuramate--alanine ligase from Acinetobacter baumannii AB5075-UW with AMPPNP
To be Published
|
|
7U0S
 
 | | Crystal Structure of FK506-binding protein 1A from Aspergillus fumigatus Bound to Ascomycin | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, ACETATE ION, FK506-binding protein 1A, ... | | Authors: | DeBouver, N.D, Fox III, D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
