7MGZ
 
 | | Human CTPS1 bound to UTP, AMPPNP, and glutamine | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH1
 
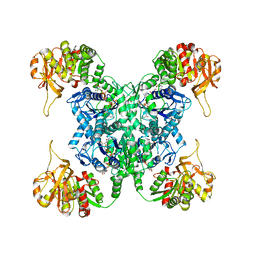 | | Human CTPS2 bound to CTP | | Descriptor: | CTP synthase 2, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIG
 
 | | Human CTPS1 bound to inhibitor T35 | | Descriptor: | 2-{2-[(cyclopropanesulfonyl)amino]-1,3-thiazol-4-yl}-2-methyl-N-{5-[6-(trifluoromethyl)pyrazin-2-yl]pyridin-2-yl}propanamide, CTP synthase 1, GLUTAMINE, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH0
 
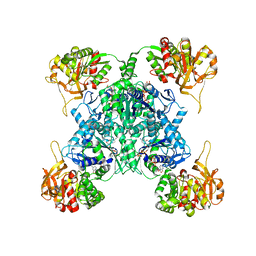 | | Human CTPS1 bound to CTP | | Descriptor: | CTP synthase 1, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIU
 
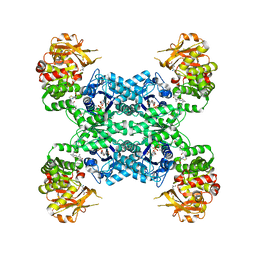 | | Mouse CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M6T
 
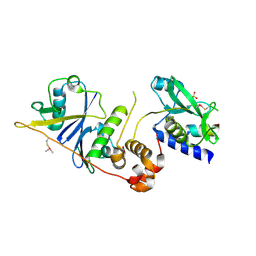 | | Crystal structure of SOCS2/ElonginB/ElonginC bound to a non-canonical peptide that enhances phospho-peptide binding | | Descriptor: | Elongin-B, Elongin-C, Non-canonical peptide F3, ... | | Authors: | Kershaw, N.J, Li, K, Linossi, E.M, Nicholson, S.E. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.
Nat Commun, 12, 2021
|
|
7MIH
 
 | | Human CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1K04
 
 | |
6UDS
 
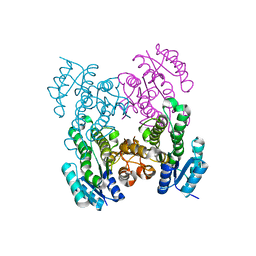 | |
1L5S
 
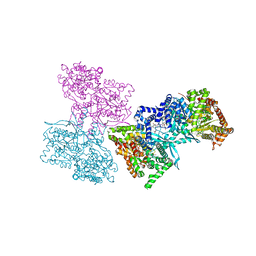 | | Human liver glycogen phosphorylase complexed with uric acid, N-Acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
6U9M
 
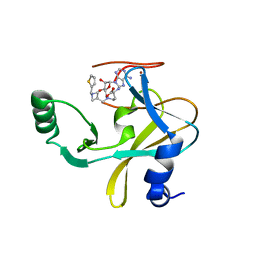 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
1L5Q
 
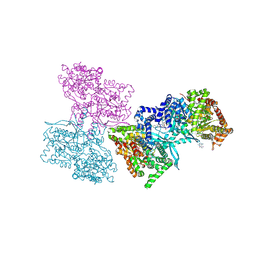 | | Human liver glycogen phosphorylase a complexed with caffeine, N-Acetyl-beta-D-glucopyranosylamine, and CP-403700 | | Descriptor: | CAFFEINE, Glycogen phosphorylase, liver form, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-07 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
6U07
 
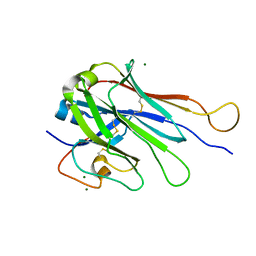 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
6UBY
 
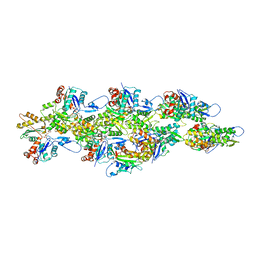 | | Isolated cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1AU7
 
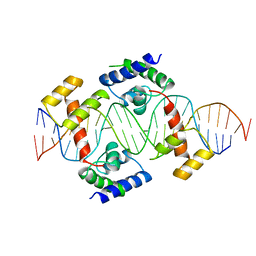 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
1E1X
 
 | |
1HNE
 
 | | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-angstroms resolution | | Descriptor: | HUMAN LEUCOCYTE ELASTASE, METHOXYSUCCINYL-ALA-ALA-PRO-ALA CHLOROMETHYL KETONE INHIBITOR | | Authors: | Navia, M.A, Mckeever, B.M, Springer, J.P, Lin, T.-Y, Williams, H.R, Fluder, E.M, Dorn, C.P, Hoogsteen, K. | | Deposit date: | 1989-04-10 | | Release date: | 1989-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-A resolution.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
1E0P
 
 | | L intermediate of bacteriorhodopsin | | Descriptor: | BACTERIORHODOPSIN, GROUND STATE, RETINAL | | Authors: | Royant, A, Edman, K, Ursby, T, Pebay-Peyroula, E, Landau, E.M, Neutze, R. | | Deposit date: | 2000-04-04 | | Release date: | 2000-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Helix Deformation is Coupled to Vectorial Proton Transport in Bacteriorhodopsin'S Photocycle
Nature, 406, 2000
|
|
1E1V
 
 | |
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1IG5
 
 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1JXH
 
 | | 4-Amino-5-hydroxymethyl-2-methylpyrimidine Phosphate Kinase from Salmonella typhimurium | | Descriptor: | PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Cheng, G, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2001-09-07 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 A resolution.
Structure, 10, 2002
|
|
1CRC
 
 | | CYTOCHROME C AT LOW IONIC STRENGTH | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Sanishvili, R, Volz, K.W, Westbrook, E.M, Margoliash, E. | | Deposit date: | 1995-03-22 | | Release date: | 1996-03-08 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The low ionic strength crystal structure of horse cytochrome c at 2.1 A resolution and comparison with its high ionic strength counterpart.
Structure, 3, 1995
|
|
1M6Z
 
 | | Crystal structure of reduced recombinant cytochrome c4 from Pseudomonas stutzeri | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome c4, GLYCEROL, ... | | Authors: | Noergaard, A, Harris, P, Larsen, S, Christensen, H.E.M. | | Deposit date: | 2002-07-18 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural comparison of recombinant Pseudomonas stutzeri cytochrome c4 in two oxidation states
To be Published
|
|
1M70
 
 | |
