2KTZ
 
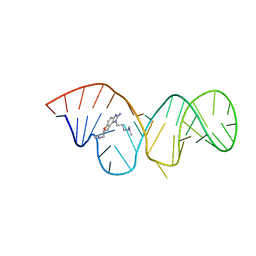 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1LH1
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | ACETATE ION, LEGHEMOGLOBIN (ACETO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
1HBS
 
 | |
2AAI
 
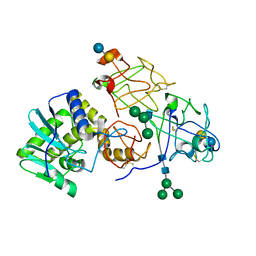 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
2PYP
 
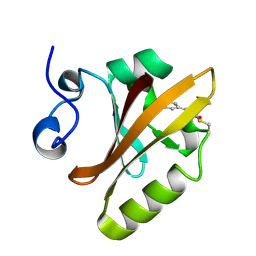 | | PHOTOACTIVE YELLOW PROTEIN, PHOTOSTATIONARY STATE, 50% GROUND STATE, 50% BLEACHED | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Borgstahl, G.E.O, Ng, K, Ren, Z, Pradervand, C, Burke, P, Srajer, V, Teng, T, Schildkamp, W, Mcree, D.E, Moffat, K, Getzoff, E.D. | | Deposit date: | 1997-02-03 | | Release date: | 1998-04-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a protein photocycle intermediate by millisecond time-resolved crystallography.
Science, 275, 1997
|
|
2POR
 
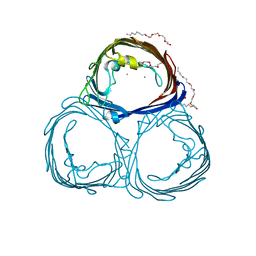 | |
2A0Q
 
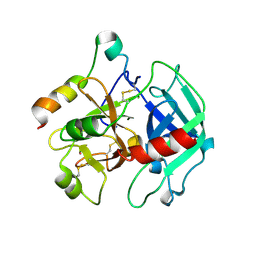 | | Structure of thrombin in 400 mM potassium chloride | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Papaconstantinou, M, Carrell, C.J, Pineda, A.O, Bobofchak, K.M, Mathews, F.S, Flordellis, C.S, Maragoudakis, M.E, Tsopanoglou, N.E, di Cera, E. | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thrombin Functions through Its RGD Sequence in a Non-canonical Conformation.
J.Biol.Chem., 280, 2005
|
|
1H8E
 
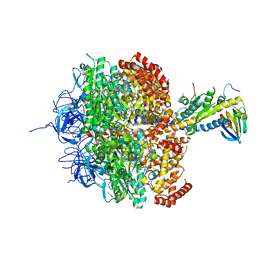 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
1GRB
 
 | |
2KU0
 
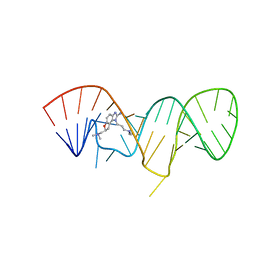 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8UWX
 
 | |
6URF
 
 | | Malic enzyme from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, NAD-dependent malic enzyme | | Authors: | Cuthbert, B.J, Burley, K.H, Goulding, C.W, Mathews, E.I, Beste, D.J. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Molecular Dynamics of Mycobacterium tuberculosis Malic Enzyme, a Potential Anti-TB Drug Target.
Acs Infect Dis., 7, 2021
|
|
8DYH
 
 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUU
 
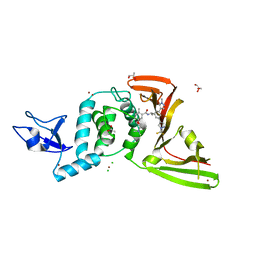 | | SARS-Cov-2 papain-like protease (PLpro) with inhibitor Jun12162 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7U5H
 
 | | Cryo-EM Structure of DNPEP | | Descriptor: | Aspartyl aminopeptidase, ZINC ION | | Authors: | Morgan, C.E, Yu, E.W, Zhang, Z. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
8UVM
 
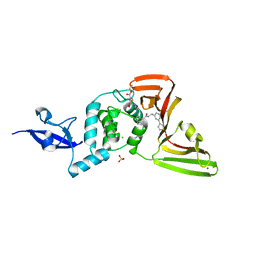 | | SARS-CoV-2 papain-like protease (PLpro) complex with covalent inhibitor Jun11313 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Ansari, A, Tan, B, Chopra, A, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7P7P
 
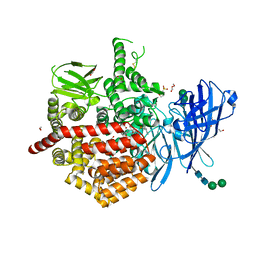 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
6DXK
 
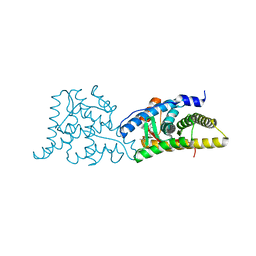 | | Glucocorticoid Receptor in complex with Compound 11 | | Descriptor: | (8S,11R,13S,14S,17S)-11-[4-(dimethylamino)phenyl]-17-(3,3-dimethylbut-1-yn-1-yl)-17-hydroxy-13-methyl-1,2,6,7,8,11,12,13,14,15,16,17-dodecahydro-3H-cyclopenta[a]phenanthren-3-one (non-preferred name), Glucocorticoid receptor | | Authors: | Rew, Y, Du, X, Eksterowicz, J, Zhou, H, Jahchan, N, Zhu, L, Yan, X, Kawai, H, McGee, L.R, Medina, J.C, Huang, T, Chen, C, Zavorotinskaya, T, Sutimantanapi, D, Waszczuk, J, Jackson, E, Huang, E, Ye, Q, Fantin, V.R, Daqing, S. | | Deposit date: | 2018-06-29 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of a Potent and Selective Steroidal Glucocorticoid Receptor Antagonist (ORIC-101).
J. Med. Chem., 61, 2018
|
|
7U5N
 
 | |
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
8UUF
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
