6Z1R
 
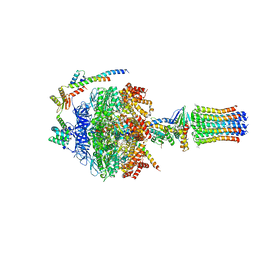 | | bovine ATP synthase F1-peripheral stalk domain, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8CHE
 
 | |
8CN9
 
 | |
8G9O
 
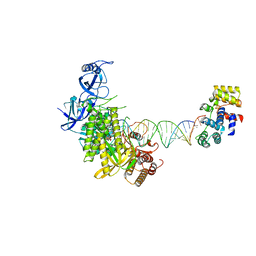 | | Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6QFA
 
 | | CryoEM structure of a beta3K279T GABA(A)R homomer in complex with histamine and megabody Mb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit beta-3, HISTAMINE, ... | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkoening, A, Zoegg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2019-01-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
7QCZ
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding canthaxanthin in the C2 space group | | Descriptor: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | Authors: | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
4TQC
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FDQ
 
 | |
7QUQ
 
 | | BtubA(R284G,K286D,F287G):BtubB bacterial tubulin M-loop mutant forming a single protofilament (Prosthecobacter dejongeii) | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Tubulin beta, Tubulin domain-containing protein | | Authors: | Wagstaff, J, Planelles-Herrero, V.J, Derivery, E, Lowe, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Diverse cytomotive actins and tubulins share a polymerization switch mechanism conferring robust dynamics.
Sci Adv, 9, 2023
|
|
6WDT
 
 | | Enterovirus D68 in complex with human monoclonal antibody EV68-228 | | Descriptor: | EV68-228 heavy chain, EV68-228 light chain, viral protein 1, ... | | Authors: | Fu, J, Vogt, M.R, Klose, T, Crowe, J.E, Rossmann, M.G, Kuhn, R.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human antibodies neutralize enterovirus D68 and protect against infection and paralytic disease.
Sci Immunol, 5, 2020
|
|
7OF5
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 and GTPBP5 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
6AN8
 
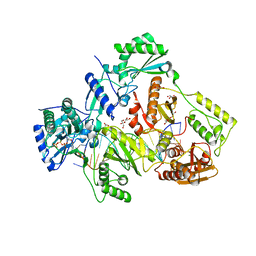 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6ASW
 
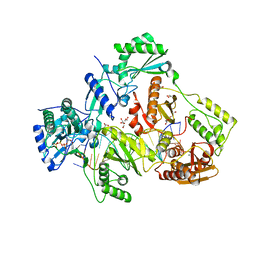 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-25 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
1C4T
 
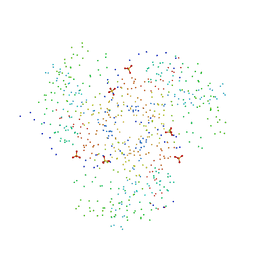 | | CATALYTIC DOMAIN FROM TRIMERIC DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | PROTEIN (DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE), SULFATE ION | | Authors: | Knapp, J.E, Carroll, D, Lawson, J.E, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1999-09-22 | | Release date: | 2000-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expression, purification, and structural analysis of the trimeric form of the catalytic domain of the Escherichia coli dihydrolipoamide succinyltransferase.
Protein Sci., 9, 2000
|
|
7OF0
 
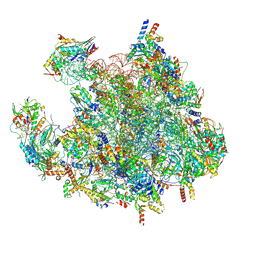 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
8G99
 
 | | Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9F
 
 | | Complete auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G9L
 
 | | DNA initiation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FIX
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8FIY
 
 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
6VZQ
 
 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6QNA
 
 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
