2VHQ
 
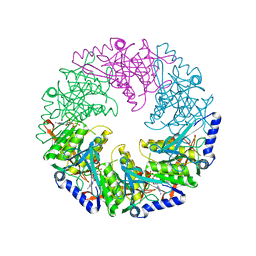 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ATP AND MG | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
4U8A
 
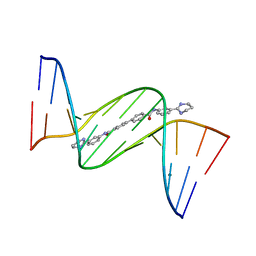 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1503 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-bis[3-(1,4,5,6-tetrahydropyrimidin-2-yl)phenyl]biphenyl-4,4'-dicarboxamide | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
6C1H
 
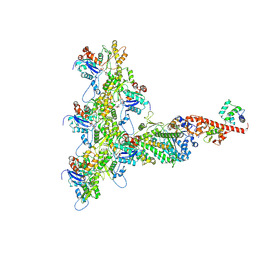 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C29
 
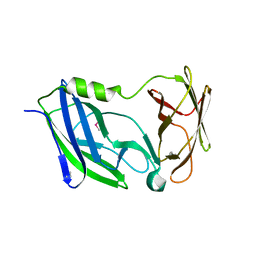 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | Descriptor: | Putative metal resistance protein | | Authors: | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | Deposit date: | 2018-01-07 | | Release date: | 2018-03-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
1OGV
 
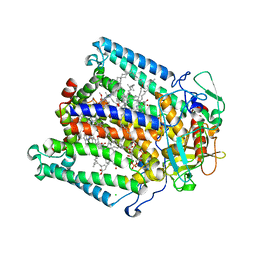 | | Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Katona, G, Andreasson, U, Landau, E.M, Andreasson, L.-E, Neutze, R. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lipidic Cubic Phase Crystal Structure of the Photosynthetic Reaction Centre from Rhodobacter Sphaeroides at 2.35 A Resolution
J.Mol.Biol., 331, 2003
|
|
8A7P
 
 | |
6NID
 
 | |
8A0G
 
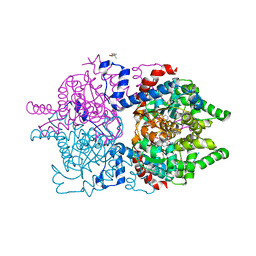 | | Human deoxyhypusine synthase with trapped transition state | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,3-DIAMINOPROPANE, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
6NJM
 
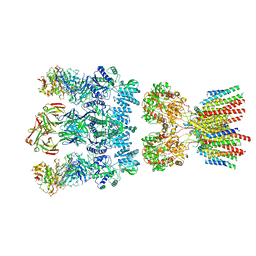 | | Architecture and subunit arrangement of native AMPA receptors | | Descriptor: | 15F1 Fab heavy chain, 15F1 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gouaux, E, Zhao, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-04-24 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM.
Science, 364, 2019
|
|
8A7Q
 
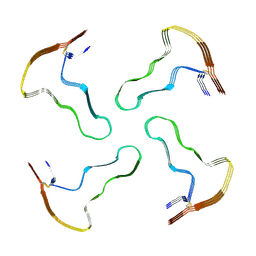 | |
7MLU
 
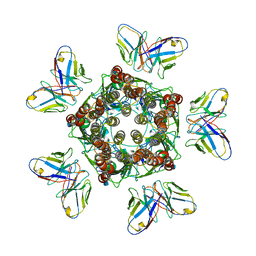 | |
5HD4
 
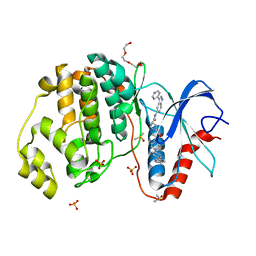 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
2V4B
 
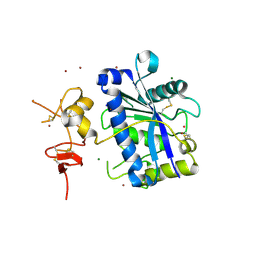 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
2OCV
 
 | | Structural basis of Na+ activation mimicry in murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
1R4Q
 
 | | Shiga toxin | | Descriptor: | SHT cytotoxin A subunit, Shigella toxin chain B | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
4H2J
 
 | |
2V90
 
 | | Crystal structure of the 3rd PDZ domain of intestine- and kidney- enriched PDZ domain IKEPP (PDZD3) | | Descriptor: | PDZ DOMAIN-CONTAINING PROTEIN 3, SULFATE ION | | Authors: | Uppenberg, J, Gileadi, C, Phillips, C, Elkins, J, Bunkoczi, G, Cooper, C, Pike, A.C.W, Salah, E, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D.A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the 3Rd Pdz Domain of Intestine- and Kidney-Enriched Pdz Domain Ikepp (Pdzd3)
To be Published
|
|
1P0Z
 
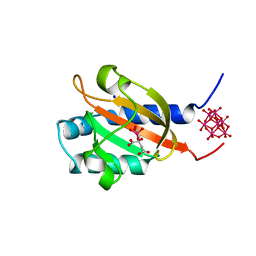 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
5KRF
 
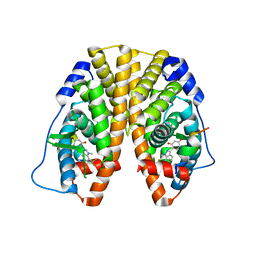 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Dynamic WAY derivative, 1a | | Descriptor: | 4-[1-methyl-7-(trifluoromethyl)indazol-3-yl]benzene-1,3-diol, Estrogen receptor, KHKILHRLLQDSSS Peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2CKM
 
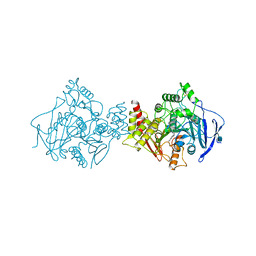 | | Torpedo californica acetylcholinesterase complexed with alkylene- linked bis-tacrine dimer (7 carbon linker) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N,N'-DI-1,2,3,4-TETRAHYDROACRIDIN-9-YLHEPTANE-1,7-DIAMINE | | Authors: | Brumshtein, B, Rydberg, E.H, Greenblatt, H.M, Wong, D.M, Shaya, D, Williams, L.D, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2006-04-20 | | Release date: | 2006-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: Binding of Bis5-tacrine produces a dramatic rearrangement in the active-site gorge.
J. Med. Chem., 49, 2006
|
|
2CEK
 
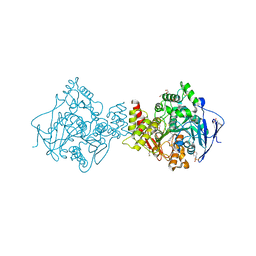 | | Conformational Flexibility in the Peripheral Site of Torpedo californica Acetylcholinesterase Revealed by the Complex Structure with a Bifunctional Inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sanson, B, Colletier, J.P, Nachon, F, Gabellieri, E, Fattorusso, C, Campiani, G, Weik, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational flexibility in the peripheral site of Torpedo californica acetylcholinesterase revealed by the complex structure with a bifunctional inhibitor.
J. Am. Chem. Soc., 128, 2006
|
|
5LAC
 
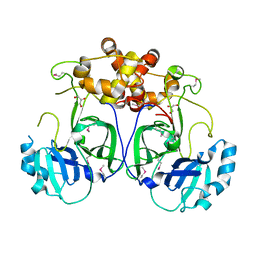 | |
6NFV
 
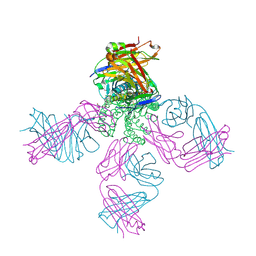 | | Structure of the KcsA-G77C mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YJ3
 
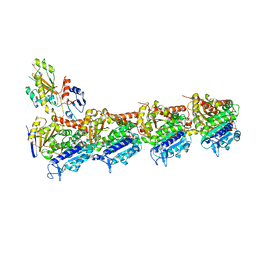 | | Crystal structure of tubulin bound to compound 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(4-ethoxyphenyl)-3-(2-methoxyphenyl)-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
