6R6Y
 
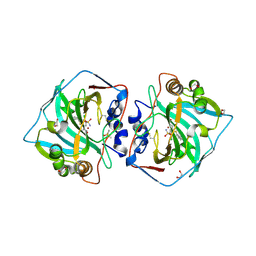 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-chloro-2-cyclohexylsulfanyl-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
5LIH
 
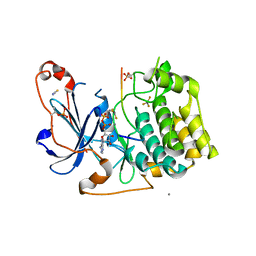 | | Structure of a peptide-substrate bound to PKCiota core kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MANGANESE (II) ION, ... | | Authors: | Soriano, E.V, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization.
Dev.Cell, 38, 2016
|
|
8G3G
 
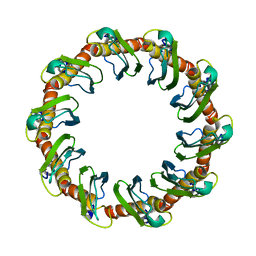 | | CryoEM structure of yeast recombination mediator Rad52 | | Descriptor: | DNA repair and recombination protein RAD52 | | Authors: | Deveryshetty, J, Basore, K, Rau, M, Fitzpatrick, J.A.J, Antony, E. | | Deposit date: | 2023-02-07 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Yeast Rad52 is a homodecamer and possesses BRCA2-like bipartite Rad51 binding modes.
Nat Commun, 14, 2023
|
|
7INS
 
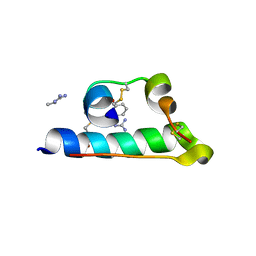 | | STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z | | Descriptor: | GENERAL PROTAMINE CHAIN, INSULIN (CHAIN A), INSULIN (CHAIN B), ... | | Authors: | Balschmidt, P, Hansen, F.B, Dodson, E, Dodson, G, Korber, F. | | Deposit date: | 1991-09-03 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of porcine insulin cocrystallized with clupeine Z.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
6R8F
 
 | | Cryo-EM structure of the Human BRISC-SHMT2 complex | | Descriptor: | BRISC and BRCA1-A complex member 2,BRCC45 (BRE, BRISC and BRCA1-A complex member 2), BRISC complex subunit Abraxas 2, ... | | Authors: | Walden, M, Hesketh, E, Tian, L, Ranson, N.A, Greenberg, R.A, Zeqiraj, E. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Metabolic control of BRISC-SHMT2 assembly regulates immune signalling.
Nature, 570, 2019
|
|
3GW6
 
 | | Intramolecular Chaperone | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6AVB
 
 | | CryoEM structure of Mical Oxidized Actin (Class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Grintsevich, E.E, Ge, P, Sawaya, M.R, Yesilyurt, H.G, Terman, J.R, Zhou, Z.H, Reisler, E. | | Deposit date: | 2017-09-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catastrophic disassembly of actin filaments via Mical-mediated oxidation.
Nat Commun, 8, 2017
|
|
5LL4
 
 | |
6OGF
 
 | | 70S termination complex with RF2 bound to the UGA codon. Partially rotated ribosome with RF2 bound (Structure III). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OI1
 
 | | Crystal structure of human WDR5 in complex with monomethyl L-arginine | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8E04
 
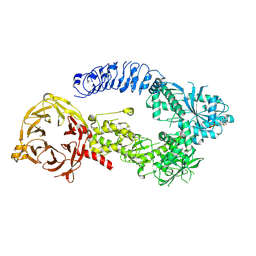 | | Structure of monomeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Mathea, S, Chatterjee, D, Knapp, S, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4Z
 
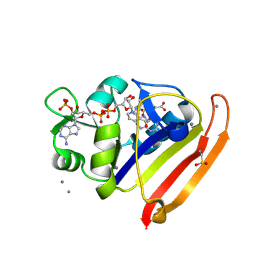 | | E. coli DHFR complex with NADP+ and folate: EF-X off model by Laue diffraction (no electric field) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8G50
 
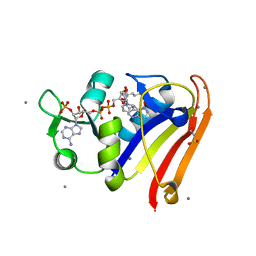 | | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3H2L
 
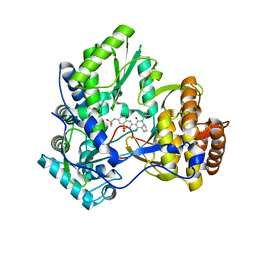 | | Crystal structure of HCV NS5B polymerase in complex with a novel bicyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(4aR,7aS)-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,4a,5,6,7,7a-hexahydro-1H-cyclopenta[b]pyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, NS5B polymerase | | Authors: | Han, Q, Showalter, R.E, Zhou, Q, Kissinger, C.R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of tricyclic 5,6-dihydro-1H-pyridin-2-ones as novel, potent, and orally bioavailable inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5M1O
 
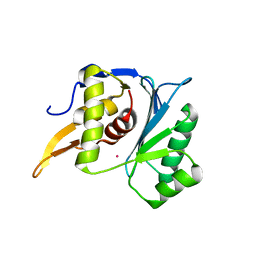 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Cobalt | | Descriptor: | COBALT (II) ION, Phage terminase large subunit | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
8OK7
 
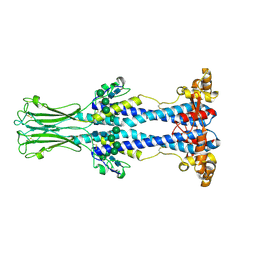 | | Variant Surface Glycoprotein VSG558 NTD | | Descriptor: | Variant surface glycoprotein 558, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, van Straaten, M, Zhong, J. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
6EZ4
 
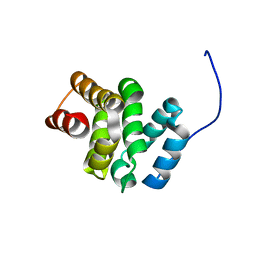 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
8GDY
 
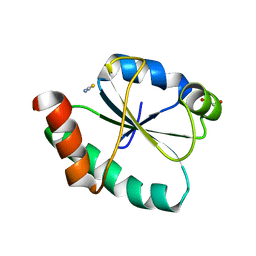 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
7JXU
 
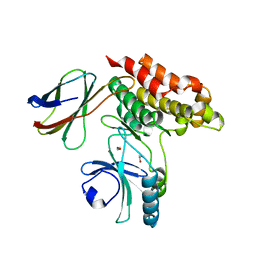 | | Structure of monobody 32 human MLKL pseudokinase domain complex | | Descriptor: | 1,2-ETHANEDIOL, Mixed lineage kinase domain-like protein, Monobody 32 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
6U9L
 
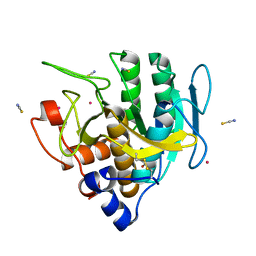 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6R71
 
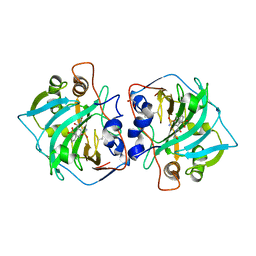 | | Crystal structure of human carbonic anhydrase isozyme XII with 2-(benzenesulfonyl)-4-chloro-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-(2-hydroxyethyl)-2-(phenylsulfonyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms.
Eur.J.Med.Chem., 185, 2020
|
|
8OFF
 
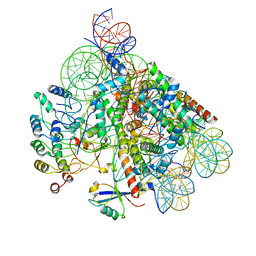 | | Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map1) | | Descriptor: | BRCA1 associated RING domain 1, DNA (142-MER), Histone H2A type 1, ... | | Authors: | Foglizzo, M, Burdett, H, Wilson, M.D, Zeqiraj, E. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BRCA1-BARD1 combines multiple chromatin recognition modules to bridge nascent nucleosomes.
Nucleic Acids Res., 51, 2023
|
|
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | Descriptor: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6UJY
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (-)-3TC-TP | | Descriptor: | Lamivudine Triphosphate, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59456086 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
