4V4S
 
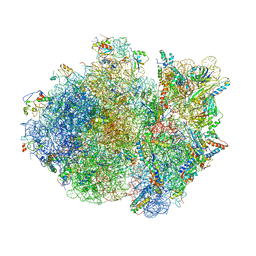 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
6Z19
 
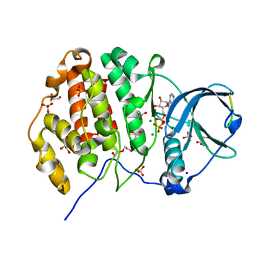 | | Crystal structure of P8C9 bound to CK2alpha | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2020-05-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
6TRG
 
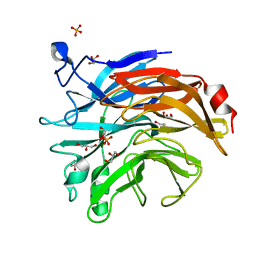 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
2E94
 
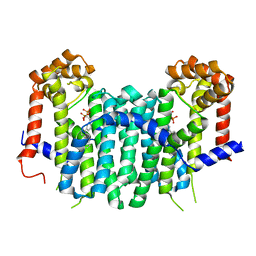 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-364 | | Descriptor: | 3-BIPHENYL-3-YL-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Ko, T.P, Cao, R, Chen, C.K.-M, Jeng, W.Y, Chang, T.H, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7RQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MTI-tripeptidyl-tRNA analog ACCA-ITM, and chloramphenicol at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MAI-tripeptidyl-tRNA analog ACCA-IAM, and chloramphenicol at 2.40A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQA
 
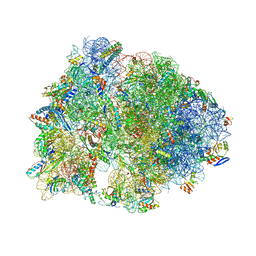 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MTI-tripeptidyl-tRNA analog ACCA-ITM at 2.40A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4NWZ
 
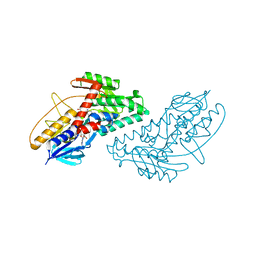 | | Structure of bacterial type II NADH dehydrogenase from Caldalkalibacillus thermarum at 2.5A resolution | | Descriptor: | FAD-dependent pyridine nucleotide-disulfide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakatani, Y, Heikal, A, Lott, J.S, Sazanov, L.A, Baker, E.N, Cook, G.M. | | Deposit date: | 2013-12-07 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation.
Mol.Microbiol., 91, 2014
|
|
4NY2
 
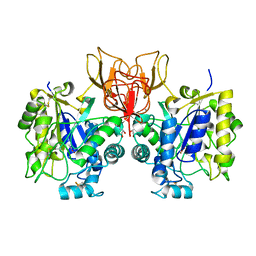 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3T60
 
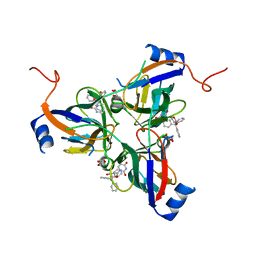 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-(tritylamino)uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
6DKW
 
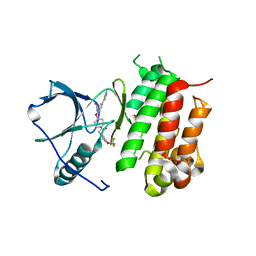 | |
7RQC
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MFI-tripeptidyl-tRNA analog ACCA-IFM at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6TQB
 
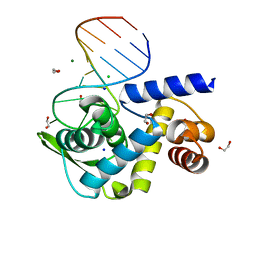 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE1 SL RNA motif | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
2E76
 
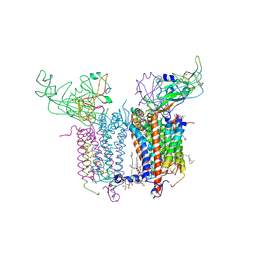 | | Crystal Structure of the Cytochrome b6f Complex with tridecyl-stigmatellin (TDS) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
6DTZ
 
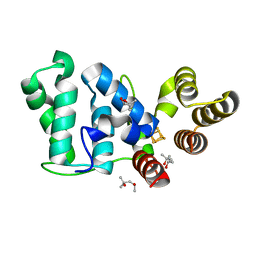 | | Crystal structure of eukaryotic DNA primase large subunit iron-sulfur cluster domain, Y397F mutant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, ... | | Authors: | Salay, L.E, Chazin, W.J. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Yeast require redox switching in DNA primase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2E95
 
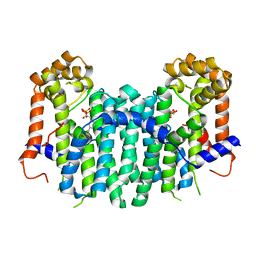 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-675 | | Descriptor: | 1-HYDROXY-2-[3'-(NAPHTHALENE-2-SULFONYLAMINO)-BIPHENYL-3-YL]ETHYLIDENE-1,1-BISPHOSPHONIC ACID, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Cao, R, Ko, T.P, Jeng, W.Y, Chang, T.H, Chen, C.K.-M, Liang, P.H, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-01-24 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4O24
 
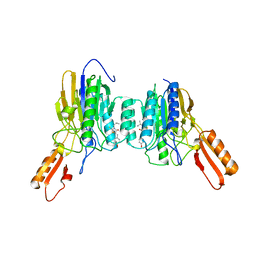 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{Z})-5-[(4-hydroxyphenyl)methylidene]-3-(2-methylpropyl)-2-sulfanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
4WCZ
 
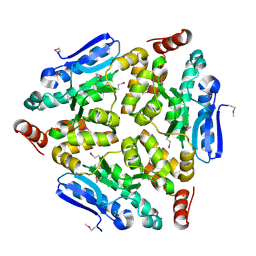 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
5HVD
 
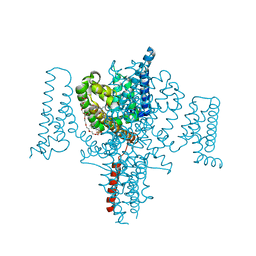 | | Full length Open-form Sodium Channel NavMs I218C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Booker, J.E, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2016-01-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The complete structure of an activated open sodium channel.
Nat Commun, 8, 2017
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
6T8X
 
 | | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, 5-(4-bromophenyl)-~{N}-(4-piperazin-1-ylphenyl)-~{N}-(pyridin-2-ylmethyl)furan-2-carboxamide, CHLORIDE ION, ... | | Authors: | Beaumont, E.J, Barker, J. | | Deposit date: | 2019-10-25 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of MAPKAPK2 (MK2) complexed with PF-3644022 and 5-(4-bromophenyl)-N-[4-(1-piperazinyl)phenyl]-N-(2-pyridinylmethyl)-2-furancarboxamide
To Be Published
|
|
7R9V
 
 | | Structure of PIK3CA with covalent inhibitor 19 | | Descriptor: | N-[2-(4-{4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethyl]-1-(prop-2-enoyl)piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Covalent Proximity Scanning of a Distal Cysteine to Target PI3K alpha.
J.Am.Chem.Soc., 144, 2022
|
|
7R9Y
 
 | | Structure of PIK3CA with covalent inhibitor 22 | | Descriptor: | N-[2-(4-{4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethyl]-N-methyl-1-(prop-2-enoyl)piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Covalent Proximity Scanning of a Distal Cysteine to Target PI3K alpha.
J.Am.Chem.Soc., 144, 2022
|
|
