1XTQ
 
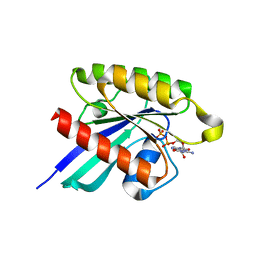 | | Structure of small GTPase human Rheb in complex with GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Yu, Y, Ding, J. | | Deposit date: | 2004-10-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Unique Biological Function of Small GTPase RHEB
J.Biol.Chem., 280, 2005
|
|
1XTS
 
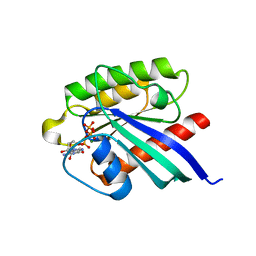 | | Structure of small GTPase human Rheb in complex with GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Yu, Y, Ding, J. | | Deposit date: | 2004-10-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Unique Biological Function of Small GTPase RHEB
J.Biol.Chem., 280, 2005
|
|
1XTR
 
 | |
1T0L
 
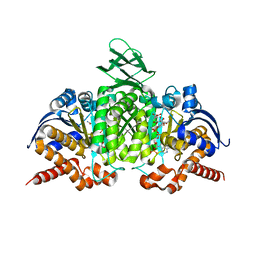 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-10 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
2DSD
 
 | |
2DSB
 
 | |
6JWP
 
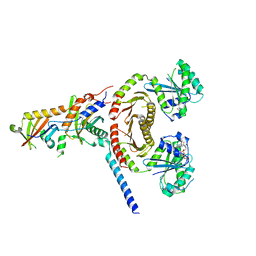 | | crystal structure of EGOC | | Descriptor: | Ego2, GTP-binding protein GTR1, GTP-binding protein GTR2, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2019-04-21 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases.
Sci Adv, 5, 2019
|
|
3KT6
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with Trp | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
6KE3
 
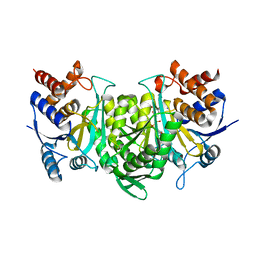 | |
3KT8
 
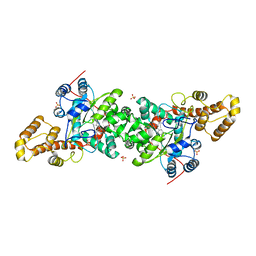 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
5C77
 
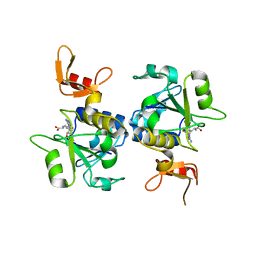 | | A novel protein arginine methyltransferase | | Descriptor: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lv, F, Zhang, T, Ding, J. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
7D0P
 
 | |
7D0Q
 
 | |
7D0O
 
 | | Crystal structure of human HBO1-BRPF2 in apo form | | Descriptor: | 1,2-ETHANEDIOL, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Li, W, Ding, J. | | Deposit date: | 2020-09-11 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | HBO1 is a versatile histone acyltransferase critical for promoter histone acylations.
Nucleic Acids Res., 49, 2021
|
|
7D0S
 
 | |
7D0R
 
 | |
3OXF
 
 | | Human lysine methyltransferase Smyd3 in complex with AdoHcy (Form I) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Xu, S, Wu, J, Sun, B, Zhong, C, Ding, J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding.
Nucleic Acids Res., 39, 2011
|
|
5Y3A
 
 | | Crystal structure of Ragulator complex (p18 49-161) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1.
Nat Commun, 8, 2017
|
|
4FG9
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-320 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
4FG8
 
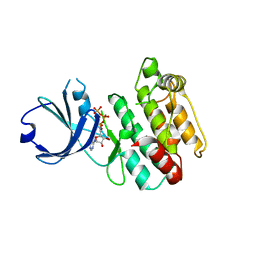 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I 1-315 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
3RPN
 
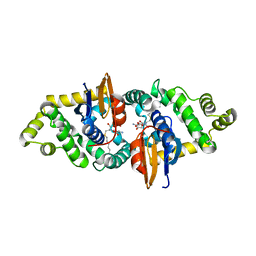 | | Crystal structure of human kappa class glutathione transferase in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase kappa 1, S-HEXYLGLUTATHIONE | | Authors: | Wang, B, Peng, Y, Zhang, T, Ding, J. | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and kinetic studies of human Kappa class glutathione transferase provide insights into the catalytic mechanism.
Biochem.J., 439, 2011
|
|
4FGB
 
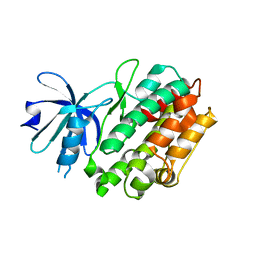 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I apo form | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
7WR3
 
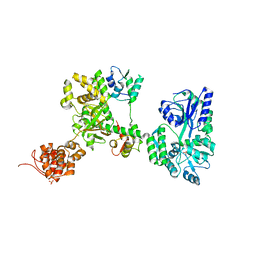 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8JKN
 
 | |
