8JPV
 
 | |
8I9I
 
 | |
7E4W
 
 | |
7ZSC
 
 | | Crystal structure of the heterodimeric human C-P4H-II with truncated alpha subunit (C-P4H-II delta281) | | Descriptor: | Prolyl 4-hydroxylase subunit alpha-2, Protein disulfide-isomerase, SULFATE ION | | Authors: | Lebedev, A, Koski, M.K, Wierenga, R.K, Murthy, A.V, Sulu, R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the collagen prolyl 4-hydroxylase (C-P4H) catalytic domain complexed with PDI: Toward a model of the C-P4H alpha 2 beta 2 tetramer.
J.Biol.Chem., 298, 2022
|
|
2C20
 
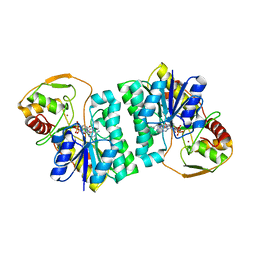 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
4MN3
 
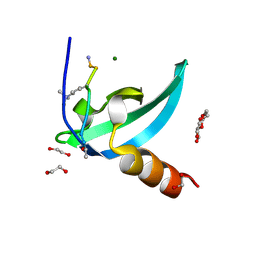 | | Chromodomain antagonists that target the polycomb-group methyllysine reader protein Chromobox homolog 7 (CBX7) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Chromobox protein homolog 7, ... | | Authors: | Chakravarthi, S, Daze, K, Douglas, S, Quon, T, Dev, A, Peng, F, Heller, M, Boulanger, M.J, Wulff, J, Hof, F. | | Deposit date: | 2013-09-09 | | Release date: | 2014-04-02 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | Chromodomain Antagonists That Target the Polycomb-Group Methyllysine Reader Protein Chromobox Homolog 7 (CBX7).
J.Med.Chem., 57, 2014
|
|
5JXX
 
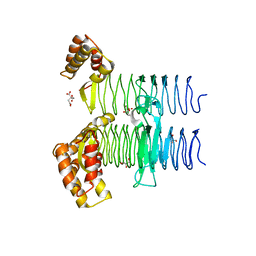 | | Crystal structure of UDP-N-acetylglucosamine O-acyltransferase (LpxA) from Moraxella catarrhalis RH4. | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION, GLYCEROL | | Authors: | Pratap, S, Kesari, P, Yadav, R, Narwal, M, Dev, A, Kumar, P. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Acyl chain preference and inhibitor identification of Moraxella catarrhalis LpxA: Insight through crystal structure and computational studies.
Int. J. Biol. Macromol., 96, 2017
|
|
5GMU
 
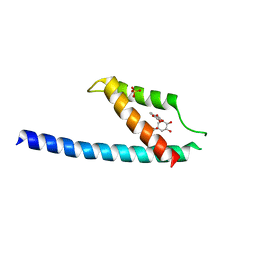 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
5GO2
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Citrate | | Descriptor: | CITRIC ACID, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
8DGE
 
 | | BoGH13ASus from Bacteroides ovatus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Alpha amylase, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
1GOV
 
 | | RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) COMPLEXED WITH SULFATE IONS | | Descriptor: | RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5T68
 
 | | Crystal structure of Syk catalytic domain in complex with a furo[3,2-d]pyrimidine | | Descriptor: | N~4~-cyclopropyl-N~2~-(3-methyl-1H-indazol-6-yl)furo[3,2-d]pyrimidine-2,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Argiriadi, M.A, Hoemann, M, Wilson, N, Banach, D, Burchat, A, Calderwood, D, Clapham, B, Cox, P, Duignan, D.B, Konopacki, D, Somal, G, Vasudevan, A. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Synthesis and optimization of furano[3,2-d]pyrimidines as selective spleen tyrosine kinase (Syk) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
4N68
 
 | | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804] | | Descriptor: | Contactin-5, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804]
to be published
|
|
4N8P
 
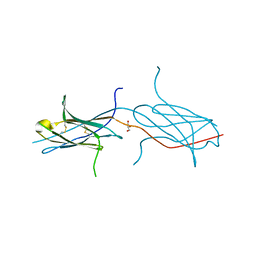 | | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Uncharacterized protein | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711]
to be published
|
|
7YLO
 
 | | Conversion of indole-3-acetic acid into indole-3-aldehyde in bacteria Metabolic network of tryptophan around the indole-3-aldehyde formation | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cheng, J, Luo, Y, Zhu, J, Tan, R, Wang, N, Lebedev, A.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Conversion of indole-3-acetic acid into indole-3-aldehyde in bacteria
Metabolic network of tryptophan around the indole-3-aldehyde formation
To Be Published
|
|
7NIT
 
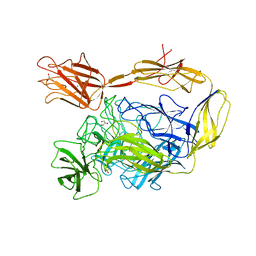 | | X-ray structure of a multidomain BbgIII from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Sanchez Rodriguez, F, Rigden, D.J, Tams, J.W, Wilting, R, Vester, J.K, Longhin, E, Krogh, K.B.R, Pache, R.A, Davies, G.J, Wilson, K.S. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6H78
 
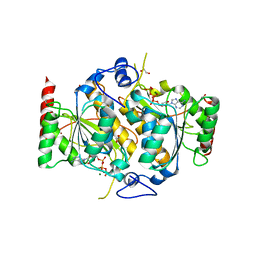 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
8FZV
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, expressed with SUMO tag | | Descriptor: | MAGNESIUM ION, Transcription factor ETV6,Anthrax toxin receptor 2, UNKNOWN ATOM OR ION | | Authors: | Pedroza Romo, M.J, Soleimani, S, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FZU
 
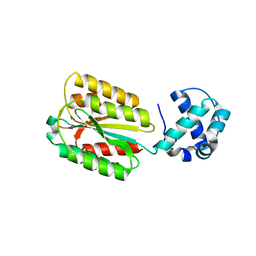 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, Expressed with SUMO tag | | Descriptor: | POTASSIUM ION, SULFATE ION, Transcription factor ETV6,Anthrax toxin receptor 2 | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
1K1H
 
 | | HETERODUPLEX OF CHIRALLY PURE METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(SMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenchova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
1K1R
 
 | | HETERODUPLEX OF CHIRALLY PURE R-METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(RMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenkova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
