7NQG
 
 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
5IXA
 
 | |
5FLG
 
 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with AMPPNP | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5HHC
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5FJZ
 
 | |
2YNO
 
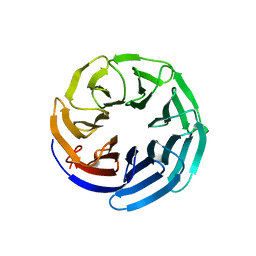 | | yeast betaprime COP 1-304H6 | | Descriptor: | COATOMER SUBUNIT BETA', POLY ALA | | Authors: | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
2Y9Z
 
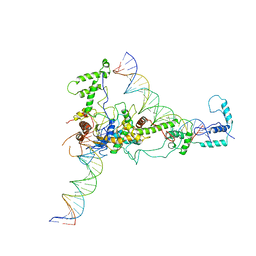 | | Chromatin Remodeling Factor ISW1a(del_ATPase) in DNA complex | | Descriptor: | I-DNA/E-DNA, IMITATION SWITCH PROTEIN 1 (DEL_ATPASE), ISWI ONE COMPLEX PROTEIN 3 | | Authors: | Yamada, K, Frouws, T.D, Angst, B, Fitzgerald, D.J, DeLuca, C, Schimmele, K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Structure and Mechanism of the Chromatin Remodelling Factor Isw1A
Nature, 472, 2011
|
|
5DDQ
 
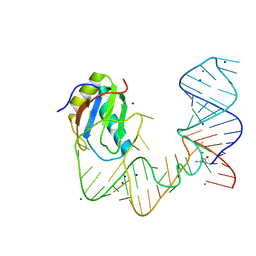 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
2Z57
 
 | | Crystal structure of G56E-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
1QH2
 
 | |
2Z56
 
 | | Crystal structure of G56S-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
5DDR
 
 | | L-glutamine riboswitch bound with L-glutamine soaked with Cs+ | | Descriptor: | CESIUM ION, GLUTAMINE, L-glutamine riboswitch RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
5DE8
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, iridium hexammine bound form. | | Descriptor: | Fragile X mental retardation protein 1, IRIDIUM HEXAMMINE ION, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1003 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5IX1
 
 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7N00
 
 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 648-1025 in complex with AUG-alpha | | Descriptor: | ALK and LTK ligand 2, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Myasnikov, A.G, Rossi, P, Miller, D.J, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
7MZY
 
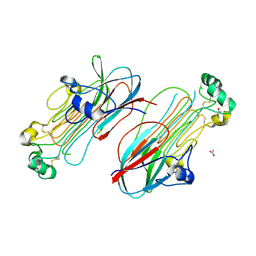 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
5EQL
 
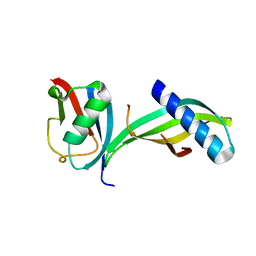 | | Isoform-specific inhibition of SUMO-dependent protein-protein interactions | | Descriptor: | SUMO-Affirmer-S2D5, Small ubiquitin-related modifier 2 | | Authors: | Hughes, D.J, Tiede, C, Hall, N, Tang, A.A.S, Trinh, C.H, Zajac, K, Mandal, U, Howell, G, Edwards, T.A, McPherson, M.J, Tomlinson, D.C, Whitehouse, A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology.
Sci Signal, 10, 2017
|
|
2Y0R
 
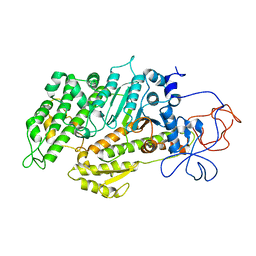 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | Descriptor: | MYOSIN-2 HEAVY CHAIN | | Authors: | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
5JG6
 
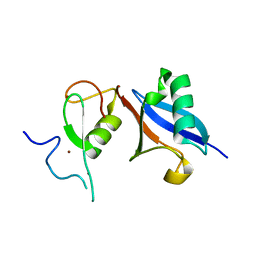 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
2B5V
 
 | | Crystal structure of glucose dehydrogenase from Haloferax mediterranei | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, glucose dehydrogenase | | Authors: | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5DKQ
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
5DOC
 
 | | Crystal structure of the Human Cytomegalovirus UL53 subunit of the NEC | | Descriptor: | GLYCEROL, Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
1PSD
 
 | |
5DKN
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | Descriptor: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
