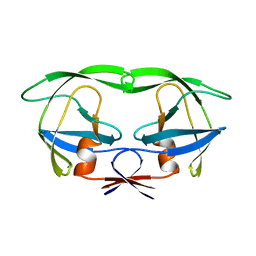
1AAQ
 
 | |
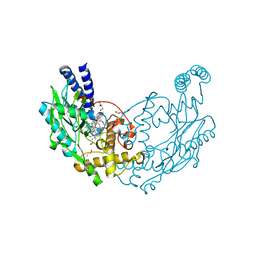
6XCX
 
 | |
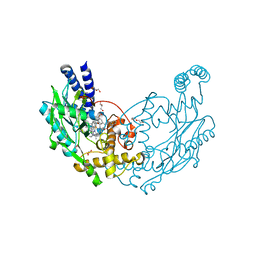
6XK8
 
 | |
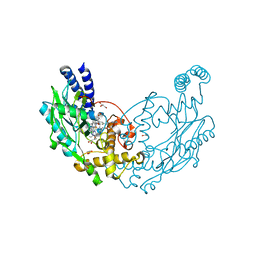
6XK7
 
 | |
6XK4
 
 | |
6XK3
 
 | |
6XK6
 
 | |
6XK5
 
 | |
6XMC
 
 | |
6DKT
 
 | |
4BV4
 
 | | Structure and allostery in Toll-Spatzle recognition | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN SPAETZLE C-106, PROTEIN TOLL, ... | | Authors: | Lewis, M.F, Arnot, C.J, Beeston, H, McCoy, A, Ashcroft, A.E, Gay, N.J, Gangloff, M. | | Deposit date: | 2013-06-24 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytokine Spatzle Binds to the Drosophila Immunoreceptor Toll with a Neurotrophin-Like Specificity and Couples Receptor Activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1ZGU
 
 | | Solution structure of the human Mms2-Ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Lewis, M.J, Saltibus, L.F, Hau, D.D, Xiao, W, Spyracopoulos, L. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Non-Covalent Interaction Between Ubiquitin and the Ubiquitin Conjugating Enzyme Variant Human MMS2.
J.Biomol.Nmr, 34, 2006
|
|
2FKD
 
 | |
2FJR
 
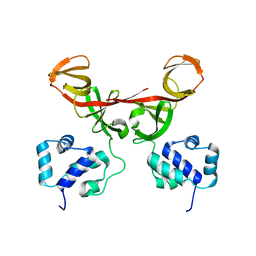 | | Crystal Structure of Bacteriophage 186 | | Descriptor: | Repressor protein CI | | Authors: | Lewis, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of cooperative regulation at an alternate genetic switch
Mol.Cell, 21, 2006
|
|
1LBG
 
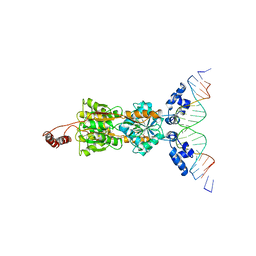 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBH
 
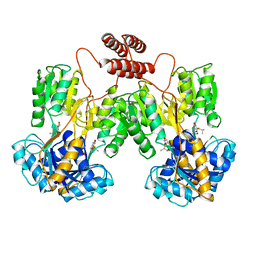 | | INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBI
 
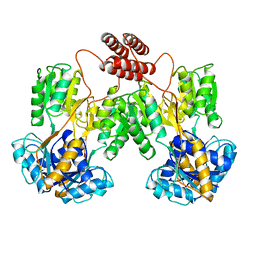 | | LAC REPRESSOR | | Descriptor: | LAC REPRESSOR | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1AFS
 
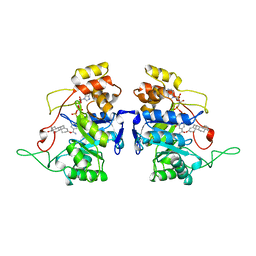 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
1EFA
 
 | |
5G6K
 
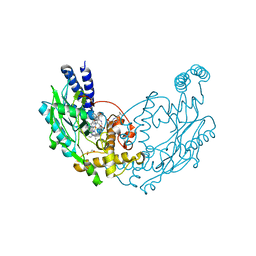 | | Structure of Bacillus subtilis Nitric Oxide Synthase I218V in complex with 7-((3-(2-(Methylamino)ethyl)phenoxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-[2-(methylamino)ethyl]phenoxy]methyl]quinolin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Lewis, M.C, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
1LRP
 
 | |
2C5I
 
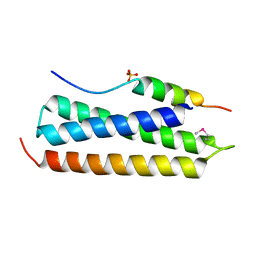 | | N-terminal domain of tlg1 complexed with N-terminus of vps51 in distorted conformation | | Descriptor: | SULFATE ION, T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1, VACUOLAR PROTEIN SORTING PROTEIN 51 | | Authors: | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | Deposit date: | 2005-10-27 | | Release date: | 2006-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
2C5J
 
 | | N-terminal domain of tlg1, domain-swapped dimer | | Descriptor: | T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1 | | Authors: | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | Deposit date: | 2005-10-27 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
2C5K
 
 | | N-terminal domain of tlg1 complexed with N-terminus of vps51 | | Descriptor: | SULFATE ION, T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1, VACUOLAR PROTEIN SORTING PROTEIN 51 | | Authors: | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | Deposit date: | 2005-10-27 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
1F39
 
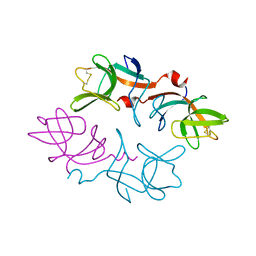 | | CRYSTAL STRUCTURE OF THE LAMBDA REPRESSOR C-TERMINAL DOMAIN | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Bell, C.E, Frescura, P, Hochschild, A, Lewis, M. | | Deposit date: | 2000-06-01 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding.
Cell(Cambridge,Mass.), 101, 2000
|
|
