6MG5
 
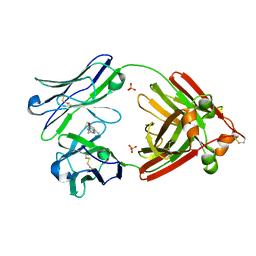 | | Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, Light chain JTO, PHOSPHATE ION | | Authors: | Morgan, G.J, Yan, N.L, Mortenson, D.E, Stanfield, R.L, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of amyloidogenic immunoglobulin light chains by small molecules.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1OYE
 
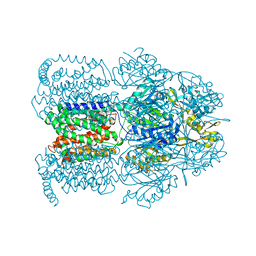 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
6MG8
 
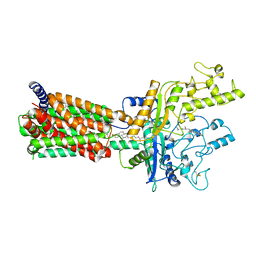 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | Descriptor: | Endoribonuclease VapD, VapX | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
1GIL
 
 | |
7KG3
 
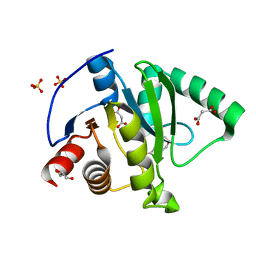 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7JYO
 
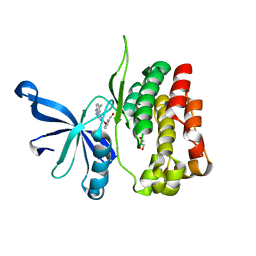 | | JAK2 JH2 in complex with JAK064 | | Descriptor: | 3-({4-amino-6-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}oxy)benzoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Puleo, D.E, Krimmer, S.G, Newton, A.S, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2020-08-31 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16127229 Å) | | Cite: | Indoloxytriazines as binding molecules for the JAK2 JH2 pseudokinase domain and its V617F variant
Tetrahedron Lett., 77, 2021
|
|
6ZPM
 
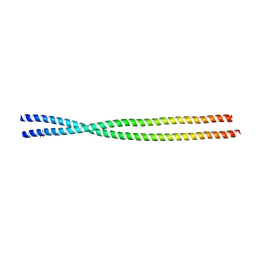 | | Crystal structure of the unconventional kinetochore protein Trypanosoma cruzi KKT4 coiled coil domain | | Descriptor: | THREONINE, Trypanosoma cruzi KKT4 117-218 | | Authors: | Ludzia, P, Lowe, D.E, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
7JYQ
 
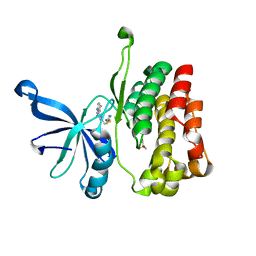 | | JAK2 JH2 in complex with JAK020 | | Descriptor: | GLYCEROL, N~2~-(4-fluorophenyl)-6-{[(5-{[(oxolan-2-yl)methyl]amino}-1,3,4-thiadiazol-2-yl)sulfanyl]methyl}-1,3,5-triazine-2,4-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Puleo, D.E, Krimmer, S.G, Newton, A.S, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2020-08-31 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85941613 Å) | | Cite: | Indoloxytriazines as binding molecules for the JAK2 JH2 pseudokinase domain and its V617F variant
Tetrahedron Lett., 77, 2021
|
|
6MPL
 
 | | Racemic M2-TM I39A crystallized from racemic detergent | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6FD1
 
 | | 7-FE FERREDOXIN FROM AZOTOBACTER VINELANDII LOW TEMPERATURE, 1.35 A | | Descriptor: | 7-FE FERREDOXIN I (FD1), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D, Stura, E.A, Mcree, D.E. | | Deposit date: | 1997-09-17 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Azotobacter vinelandii 7Fe ferredoxin at 1.35 A resolution and determination of the [Fe-S] bonds with 0.01 A accuracy.
J.Mol.Biol., 278, 1998
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6XJK
 
 | | JAK2 JH2 in complex with JAK067 | | Descriptor: | 4-({4-amino-6-[(1H-indol-5-yl)oxy]-1,3,5-triazin-2-yl}amino)benzene-1-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Puleo, D.E, Krimmer, S.G, Newton, A.S, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2020-06-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.023508 Å) | | Cite: | Explicit Representation of Cation-pi Interactions in Force Fields with 1/r4 Nonbonded Terms.
J Chem Theory Comput, 16, 2020
|
|
6X25
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7KXB
 
 | | Crystal structure of SARS-CoV-2 Nsp3 Macrodomain complex with PARG329 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]propyl}-N'-[2-(morpholin-4-yl)ethyl]thiourea, Non-structural protein 3, ... | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6DRW
 
 | |
6DRE
 
 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
7KG1
 
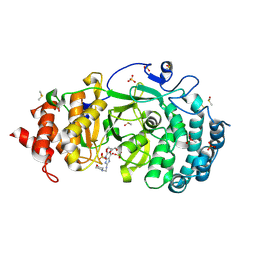 | | Structure of human PARG complexed with PARG-002 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]amino}-3,7-dihydro-1H-purine-2,6-dione, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG7
 
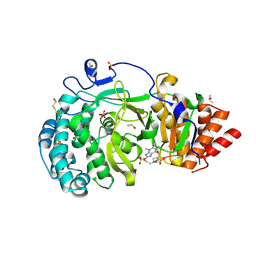 | | Structure of human PARG complexed with PARG-292 | | Descriptor: | 8-{[2-(1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)ethyl]sulfanyl}-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KFP
 
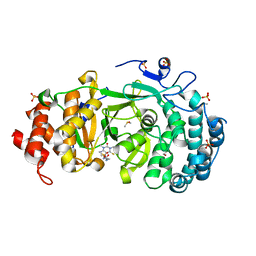 | | Structure of human PARG complexed with PARG-119 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{[2-(1,3-dimethyl-2-oxo-6-sulfanylidene-1,2,3,6-tetrahydro-7H-purin-7-yl)ethyl]carbamoyl}methanesulfonamide, ... | | Authors: | Brosey, C.A, Bommagani, S, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG0
 
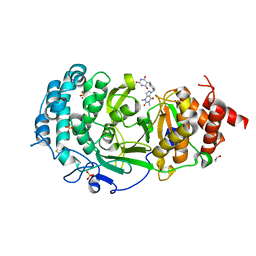 | | Structure of human PARG complexed with PARG-131 | | Descriptor: | 1,2-ETHANEDIOL, 5-({4-[(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl]phenyl}methyl)pyrimidine-2,4,6(1H,3H,5H)-trione, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Arvai, A, Bommagani, S, Brosey, C.A, Jones, D.E, Warden, L.S, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG6
 
 | | Structure of human PARG complexed with PARG-322 | | Descriptor: | 1-{2-[(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)sulfanyl]ethyl}piperidine-4-carboxylic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG8
 
 | | Structure of human PARG complexed with PARG-061 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)-2-oxoethyl]sulfanyl}-6-sulfanylidene-1,3,6,7-tetrahydro-2H-purin-2-one, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6MPN
 
 | | Racemic M2-TM I42E crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
