4H9R
 
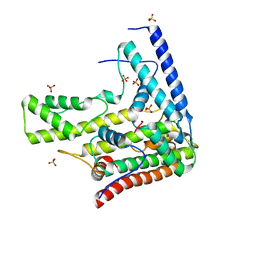 | | Complex structure 5 of DAXX(E225A)/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4GXU
 
 | |
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4F87
 
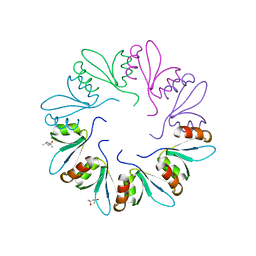 | | X-ray Crystal Structure of PlyCB | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | McGowan, S, Buckle, A.M, Fischetti, V.A, Nelson, D.C, Whisstock, J.C. | | Deposit date: | 2012-05-17 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of the streptococcal specific phage lysin PlyC.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4OAG
 
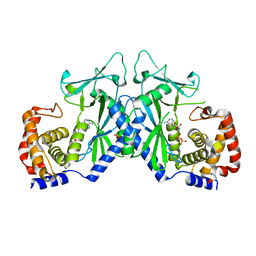 | |
4FQV
 
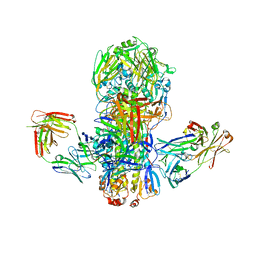 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4GMS
 
 | | Crystal structure of heterosubtypic Fab S139/1 in complex with influenza A H3 hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S139/1 heavy chain, ... | | Authors: | Lee, P.S, Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G04
 
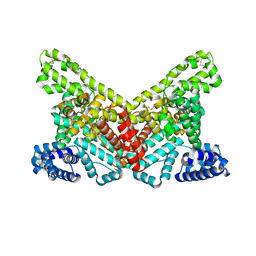 | |
4H9Q
 
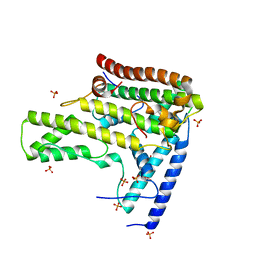 | | Complex structure 4 of DAXX(E225A)/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4PR6
 
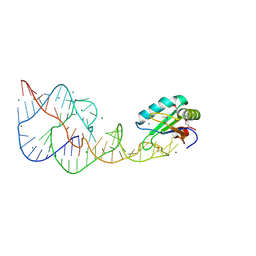 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4GXV
 
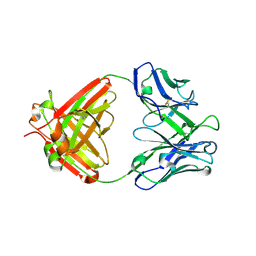 | | Crystal structure of anti-influenza virus antibody 1F1 | | Descriptor: | Antibody 1F1, heavy chain, light chain | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Influenza Human Monoclonal Antibody 1F1 Interacts with Three Major Antigenic Sites and Residues Mediating Human Receptor Specificity in H1N1 Viruses.
Plos Pathog., 8, 2012
|
|
4OAI
 
 | |
4OAF
 
 | |
4P5O
 
 | |
4G5X
 
 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G8C
 
 | |
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O6P
 
 | | Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
4G8D
 
 | |
4NPE
 
 | | High-resolution structure of C domain of staphylococcal protein A at room temperature | | Descriptor: | Immunoglobulin G-binding protein A, THIOCYANATE ION, ZINC ION | | Authors: | Deis, L.N, Pemble IV, C.W, Oas, T.G, Richardson, J.S, Richardson, D.C. | | Deposit date: | 2013-11-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Multiscale conformational heterogeneity in staphylococcal protein a: possible determinant of functional plasticity.
Structure, 22, 2014
|
|
4NXR
 
 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Neurexin-1 Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), Neurexin-2-beta Peptide, ... | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
4GGL
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-08-06 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HWA
 
 | |
4HZ0
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 7-(1H-imidazol-1-yl)-2-(pyridin-3-yl)[1,3]thiazolo[5,4-d]pyrimidin-5-amine, DNA topoisomerase 4 subunit B, MAGNESIUM ION | | Authors: | Bensen, D.C, Trzoss, M, Tari, L.W. | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
