5I4H
 
 | |
5I91
 
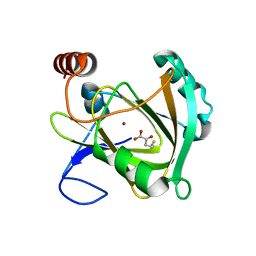 | | Structure of Mouse Acirecutone dioxygenase with to Ni2+ and 2-keto-4-(methylthio)-butyric acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, NICKEL (II) ION | | Authors: | Deshpande, A.R, Robinson, H, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
7Q0O
 
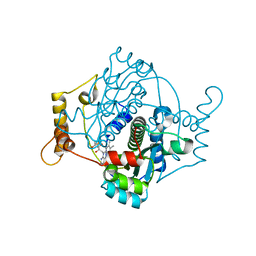 | | E. coli NfsA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | Authors: | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
6I1Z
 
 | |
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
4UML
 
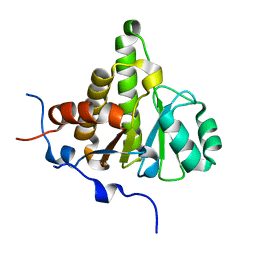 | | Crystal structure of ganglioside induced differentiation associated protein 2 (GDAP2) macro domain | | Descriptor: | GANGLIOSIDE-INDUCED DIFFERENTIATION-ASSOCIATED PROTEIN 2 | | Authors: | Elkins, J.M, Wang, J, Kopec, J, Wang, D, Strain-Damerell, C, Shrestha, L, Sieg, C, Tallant, C, Newman, J.A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2014-05-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Gdap2
To be Published
|
|
5IEV
 
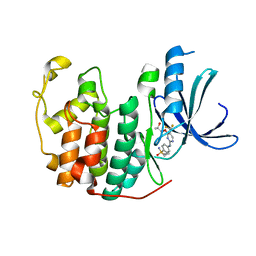 | | Crystal structure of BAY 1000394 (Roniciclib) bound to CDK2 | | Descriptor: | Cyclin-dependent kinase 2, Roniciclib | | Authors: | Ayaz, P, Andres, D, Kwiatkowski, D.A, Kolbe, C, Lienau, P, Siemeister, G, Luecking, U, Stegmann, C.M. | | Deposit date: | 2016-02-25 | | Release date: | 2016-04-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Adaption May Explain the Slow Dissociation Kinetics of Roniciclib (BAY 1000394), a Type I CDK Inhibitor with Kinetic Selectivity for CDK2 and CDK9.
Acs Chem.Biol., 11, 2016
|
|
6I4B
 
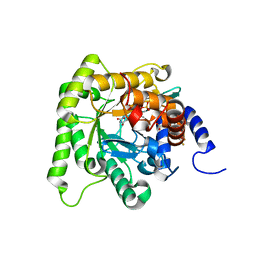 | | Plasmodium falciparum dihydroorotate dehydrogenase (DHODH) co-crystallized with 3-Hydroxy-1-methyl-5-((3-(trifluoromethyl)phenoxy)methyl)-1H-pyrazole-4-carboxylic acid | | Descriptor: | 1-methyl-3-oxidanyl-5-[[3-(trifluoromethyl)phenoxy]methyl]pyrazole-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Goyal, P, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S. | | Deposit date: | 2018-11-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Eur J Med Chem, 163, 2018
|
|
7C7Q
 
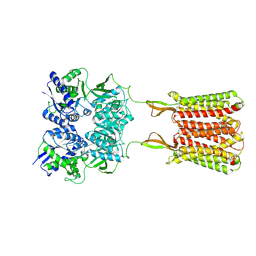 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
5I0L
 
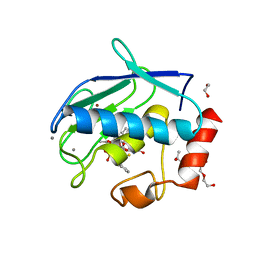 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated arylsulfonamide carboxylate water-soluble inhibitor (DC27). | | Descriptor: | (2R)-2-[{(E)-2-[({(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-bis(acetyloxy)-6-[(acetyloxy)methyl]tetrahydro-2H-pyran-2-yl}carbamothioyl)amino]ethenyl}(biphenyl-4-ylsulfonyl)amino]-3-methylbutanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
6I7I
 
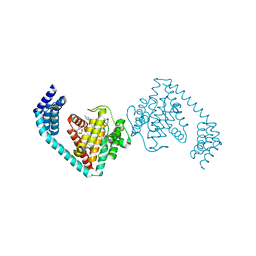 | | Crystal structure of dimeric FICD mutant K256A complexed with MgATP | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Adenosine monophosphate-protein transferase FICD, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6RZ8
 
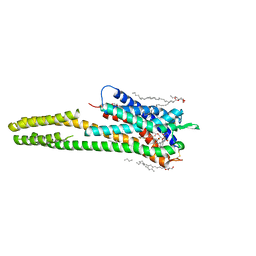 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2080365 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-[2,3-bis(fluoranyl)phenoxy]butoxy]-2-fluoranyl-phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-but yl)-2,3-dihydro-1,4-benzoxazine-2-carboxylic acid, Cysteinyl leukotriene receptor 2,Soluble cytochrome b562,Cysteinyl leukotriene receptor 2, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
7PFL
 
 | | The SARS-CoV2 major protease (Mpro) apo structure to 1.8 A resolution | | Descriptor: | Replicase polyprotein 1ab, SODIUM ION | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
1WSB
 
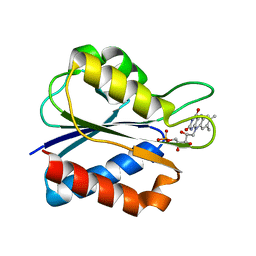 | | Flavodoxin mutant- S64C | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Marchini, N, Meneghetti, F, Rossi, G.L, Cavazzini, D, Gilardi, G. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redox properties and crystal structures of a Desulfovibrio vulgaris flavodoxin mutant in the monomeric and homodimeric forms.
Biochim.Biophys.Acta, 1794, 2009
|
|
5I8S
 
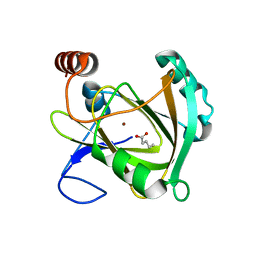 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and pentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, NICKEL (II) ION, PENTANOIC ACID | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
5I69
 
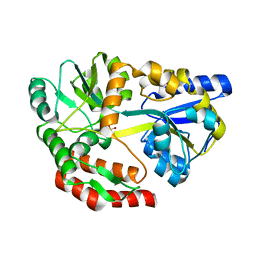 | | MBP-MamC magnetite-interaction component mutant-D70A | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Tercedor, C.V, Kolusheva, S, Gonzalez, T.P, Widdrat, M, Grimberg, N, Levi, H, Nelkenbaum, O, Davidove, G, Faivre, D, Jimenez-Lopez, C, Zarivach, R. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC.
J.Struct.Biol., 194, 2016
|
|
7PFM
 
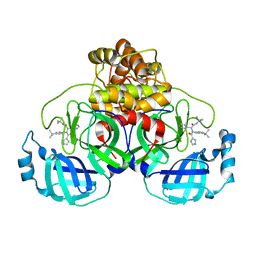 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
6I2K
 
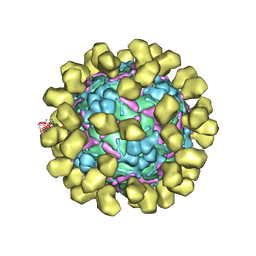 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
1WWC
 
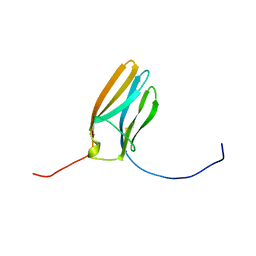 | | NT3 BINDING DOMAIN OF HUMAN TRKC RECEPTOR | | Descriptor: | PROTEIN (NT-3 GROWTH FACTOR RECEPTOR TRKC) | | Authors: | Ultsch, M.H, Wiesmann, C, Simmons, L.C, Henrich, J, Yang, M, Reilly, D, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-30 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
6I2V
 
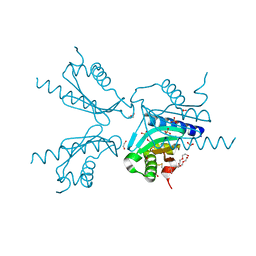 | | Pilotin from Vibrio vulnificus type 2 secretion system, EpsS. | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Howard, S.P, Estrozi, L, Bertrand, Q, Contreras-Martel, C, Strozen, T, Job, V, Martins, A, Fenel, D, Schoehn, G, Dessen, A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
1IXS
 
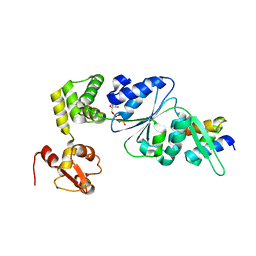 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
6I5F
 
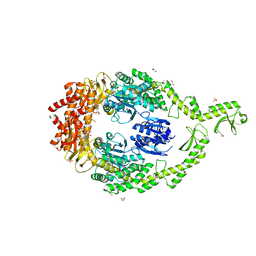 | | Crystal structure of DNA-free E.coli MutS P839E dimer mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, GLYCEROL, ... | | Authors: | Bhairosing-Kok, D, Groothuizen, F.S, Fish, A, Dharadhar, S, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2018-11-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sharp kinking of a coiled-coil in MutS allows DNA binding and release.
Nucleic Acids Res., 47, 2019
|
|
3PPD
 
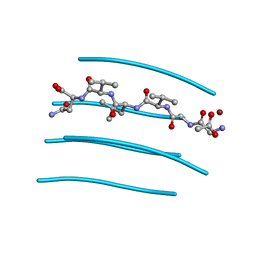 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
1IY2
 
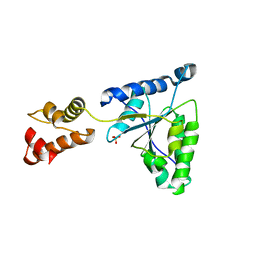 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IYT
 
 | | Solution structure of the Alzheimer's disease amyloid beta-peptide (1-42) | | Descriptor: | Alzheimer's disease amyloid | | Authors: | Crescenzi, O, Tomaselli, S, Guerrini, R, Salvadori, S, D'Ursi, A.M, Temussi, P.A, Picone, D. | | Deposit date: | 2002-09-06 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain.
EUR.J.BIOCHEM., 269, 2002
|
|
