6FM1
 
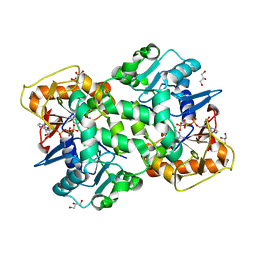 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATPanddGMP at 2.3 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
3J9G
 
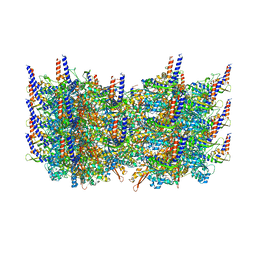 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
6T1M
 
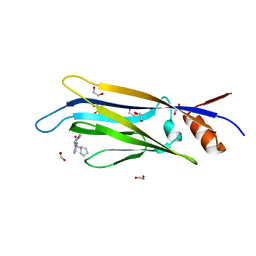 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6FNC
 
 | |
3J68
 
 | | Structural mechanism of the dynein powerstroke (pre-powerstroke state) | | Descriptor: | Dynein motor domain | | Authors: | Lin, J, Okada, K, Raytchev, M, Smith, M.C, Nicastro, D. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (30 Å) | | Cite: | Structural mechanism of the dynein power stroke.
Nat.Cell Biol., 16, 2014
|
|
6FNL
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase | | Descriptor: | Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6T2B
 
 | | Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl hydrolase family 109 protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chaberski, E.K, Fredslund, F, Teze, D, Shuoker, B, Kunstmann, S, Karlsson, E.N, Hachem, M.A, Welner, D.H. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Catalytic Acid-Base in GH109 Resides in a Conserved GGHGG Loop and Allows for Comparable alpha-Retaining and beta-Inverting Activity in an N-Acetylgalactosaminidase from Akkermansia muciniphila
Acs Catalysis, 2020
|
|
3JBS
 
 | |
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
6FP7
 
 | | mTFP1/DARPin 1238_E11 complex in space group P6522 | | Descriptor: | DARPin1238_E11, GFP-like fluorescent chromoprotein cFP484, GLYCEROL | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
6FPK
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FWT
 
 | | Crystal structure of human wild type beta-1,4-galactosyltransferase-1 (B4GalT1) in apo-open monomeric form | | Descriptor: | Beta-1,4-galactosyltransferase 1, GLYCEROL | | Authors: | Harrus, D, Kellokumpu, S, Glumoff, T. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | The dimeric structure of wild-type human glycosyltransferase B4GalT1.
PLoS ONE, 13, 2018
|
|
6FQE
 
 | | Phosphotriesterase PTE_A53_4 | | Descriptor: | (4~{S},6~{R})-2,2,6-trimethyl-1,3-dioxan-4-ol, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
6FQK
 
 | | GluA2(flop) S729C ligand binding core dimer bound to ZK200775 at 1.98 Angstrom resolution | | Descriptor: | Glutamate receptor 2,Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.98010445 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6T3B
 
 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
3JUZ
 
 | |
3J1V
 
 | |
6FP8
 
 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FQ3
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6SZY
 
 | | Crystal structure of YTHDC1 with fragment 12 (DHU_DC1_150) | | Descriptor: | 4-propan-2-yl-5,6,7,8-tetrahydro-4~{a}~{H}-quinazolin-2-one, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T04
 
 | | Crystal structure of YTHDC1 with fragment 17 (DHU_DC1_042) | | Descriptor: | 2-phenyl-4,5-dihydro-1~{H}-imidazole, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3J9W
 
 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | Authors: | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
6T12
 
 | | Crystal structure of YTHDC1 with fragment 30 (DHU_DC1_220) | | Descriptor: | SULFATE ION, YTHDC1, ~{N},2,3-trimethyl-1~{H}-indole-5-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6FN9
 
 | |
