4PQ8
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
1MVY
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MZ0
 
 | | STRUCTURE OF MYOGLOBIN MB-YQR 316 ns AFTER PHOTOLYSIS OF CARBON MONOXIDE SOLVED FROM LAUE DATA AT RT. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N04
 
 | | Diferric chicken serum transferrin at 2.8 A resolution. | | Descriptor: | CARBONATE ION, FE (III) ION, beta-L-fucopyranose, ... | | Authors: | Guha Thakurta, P, Choudhury, D, Dasgupta, R, Dattagupta, J.K. | | Deposit date: | 2002-10-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of diferric hen serum transferrin at 2.8 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N1J
 
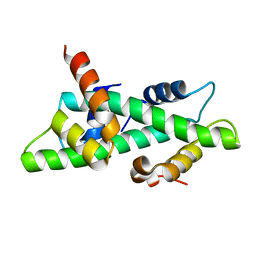 | | Crystal structure of the NF-YB/NF-YC histone pair | | Descriptor: | NF-YB, NF-YC | | Authors: | Romier, C, Cocchiarella, F, Mantovani, R, Moras, D. | | Deposit date: | 2002-10-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y
J.Biol.Chem., 278, 2003
|
|
1N29
 
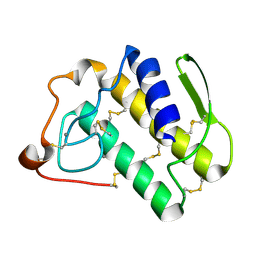 | | Crystal structure of the N1A mutant of human group IIA phospholipase A2 | | Descriptor: | CALCIUM ION, Phospholipase A2, membrane associated | | Authors: | Edwards, S.H, Thompson, D, Baker, S.F, Wood, S.P, Wilton, D.C. | | Deposit date: | 2002-10-22 | | Release date: | 2003-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism
Biochemistry, 41, 2002
|
|
1N2E
 
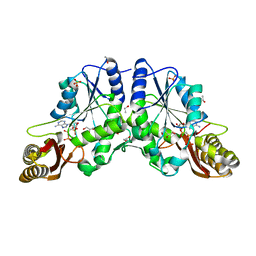 | |
4PTI
 
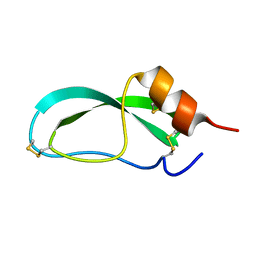 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
1N2B
 
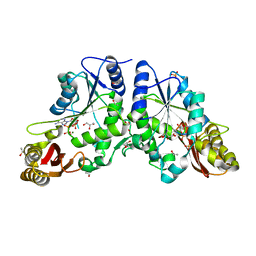 | | Crystal Structure of a Pantothenate Synthetase from M. tuberculosis in complex with AMPCPP and pantoate, higher occupancy of pantoate and lower occupancy of AMPCPP in subunit A | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ETHANOL, GLYCEROL, ... | | Authors: | Wang, S, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-22 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a pantothenate
synthetase from M. tuberculosis and its
complexes with substrates and a
reaction intermediate
Protein Sci., 12, 2003
|
|
1N2O
 
 | |
1MV9
 
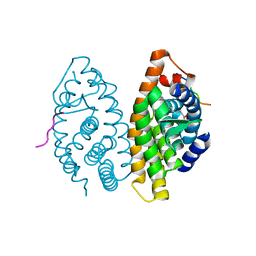 | | Crystal Structure of the human RXR alpha ligand binding domain bound to the eicosanoid DHA (Docosa Hexaenoic Acid) and a coactivator peptide | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-24 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
1MBU
 
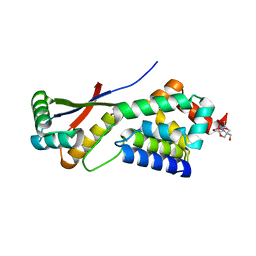 | | Crystal Structure Analysis of ClpSN heterodimer | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
4QHK
 
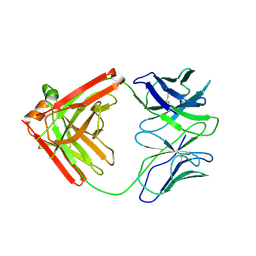 | | UCA (unbound) from CH103 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7KBB
 
 | |
7KBA
 
 | |
1M54
 
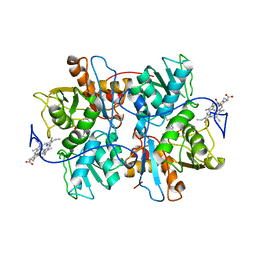 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
5VL4
 
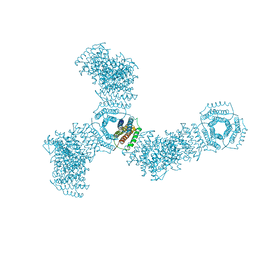 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
3WIS
 
 | | Crystal structure of Burkholderia xenovorans DmrB in complex with FMN: A Cubic Protein Cage for Redox Transfer | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | Authors: | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | Deposit date: | 2013-09-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
7MYL
 
 | |
1MNN
 
 | | Structure of the sporulation specific transcription factor Ndt80 bound to DNA | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Stuart, D, Tsang, R, Wu, C, Glover, J.N. | | Deposit date: | 2002-09-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the sporulation-specific transcription factor Ndt80 bound to DNA
Embo J., 21, 2002
|
|
7MYM
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
1MI1
 
 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
4A8E
 
 | | The structure of a dimeric Xer recombinase from archaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROBABLE TYROSINE RECOMBINASE XERC-LIKE, ... | | Authors: | Brooks, M.A, ElArnaout, T, Duranda, D, Lisboa, J, Lazar, N, Raynal, B, vanTilbeurgh, H, Serre, M, Quevillon-Cheruel, S. | | Deposit date: | 2011-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Carboxy-Terminal Alpha N Helix of the Archaeal Xera Tyrosine Recombinase is a Molecular Switch to Control Site-Specific Recombination.
Plos One, 8, 2013
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
7NAE
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
