4UW9
 
 | |
4CMR
 
 | | The crystal structure of novel exo-type maltose-forming amylase(Py04_0872) from Pyrococcus sp. ST04 | | Descriptor: | GLYCOSYL HYDROLASE/DEACETYLASE FAMILY PROTEIN | | Authors: | Park, K.-H, Jung, J.-H, Park, C.-S, Woo, E.-J. | | Deposit date: | 2014-01-17 | | Release date: | 2014-10-22 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Features Underlying the Selective Cleavage of a Novel Exo-Type Maltose-Forming Amylase from Pyrococcus Sp. St04
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5I2Q
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5I2O
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5I2L
 
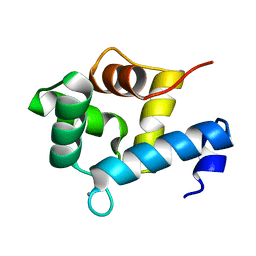 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
4BBW
 
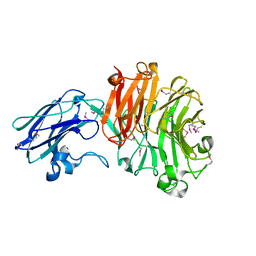 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
5H0P
 
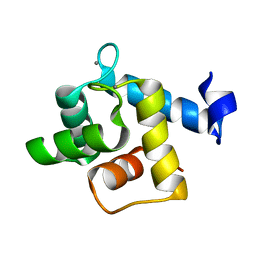 | | Crystal structure of EF-hand protein mutant | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, An, J.Y, Kang, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Mun, S.A, Jun, C.D, Song, W.K, Eom, S.H. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural mechanism underlying regulation of human EFhd2/Swiprosin-1 actin-bundling activity by Ser183 phosphorylation.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
7T7M
 
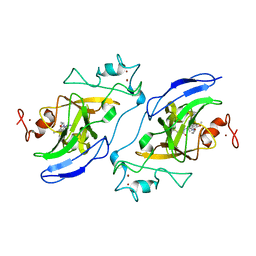 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7T7L
 
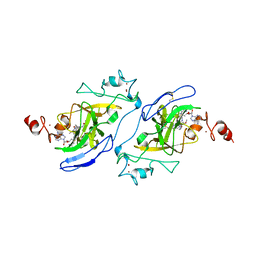 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
7CVT
 
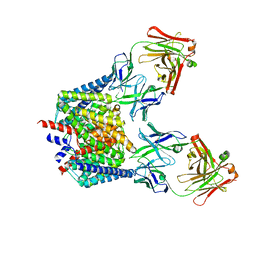 | | Crystal structure of the C85A/L194A/H234C mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
7CVS
 
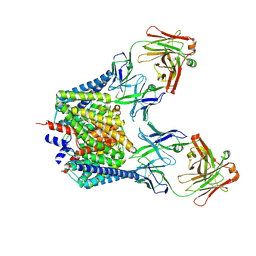 | | Crystal structure of the C85A/L194A mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
4MM2
 
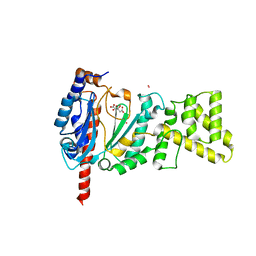 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
1K64
 
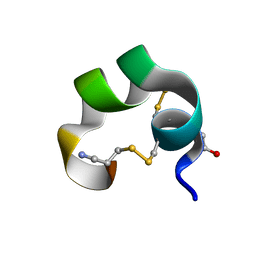 | | NMR Structue of alpha-conotoxin EI | | Descriptor: | alpha-conotoxin EI | | Authors: | Park, K.H, Suk, J.E, Jacobsen, R, Gray, W.R, McIntosh, J.M, Han, K.H. | | Deposit date: | 2001-10-15 | | Release date: | 2003-09-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin EI, a neuromuscular toxin specific for the alpha 1/delta subunit interface of torpedo nicotinic acetylcholine receptor
J.BIOL.CHEM., 276, 2001
|
|
6IY8
 
 | | DmpR-phenol complex of Pseudomonas putida | | Descriptor: | PHENOL, Positive regulator CapR, ZINC ION | | Authors: | Park, K.H, Woo, E.J. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Tetrameric architecture of an active phenol-bound form of the AAA+transcriptional regulator DmpR.
Nat Commun, 11, 2020
|
|
4MHQ
 
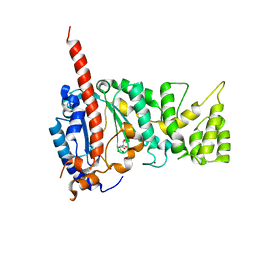 | | Crystal structure of human primase catalytic subunit | | Descriptor: | CITRIC ACID, DNA primase small subunit, ZINC ION | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human primase catalytic subunit
To be Published
|
|
6K5D
 
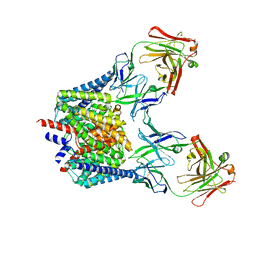 | |
6LYF
 
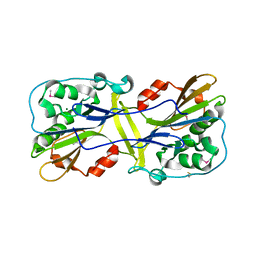 | |
6K5I
 
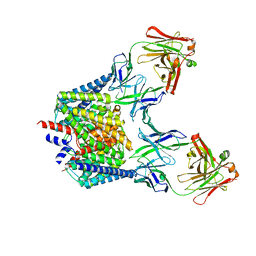 | |
6K5F
 
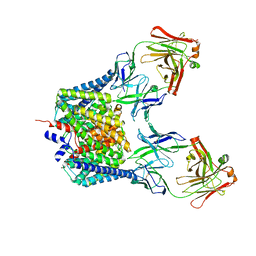 | | Crystal structure of the CLC-ec1 deltaNC in presence of 200 mM NaBr | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Park, K, Lim, H.H. | | Deposit date: | 2019-05-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K5A
 
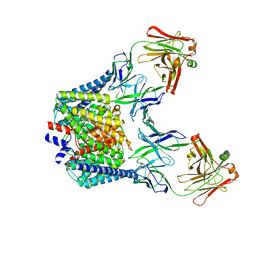 | |
6LDN
 
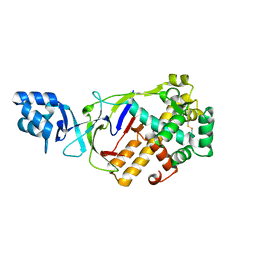 | |
6M3T
 
 | |
6M3F
 
 | |
6M3U
 
 | |
4UW2
 
 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
