9CYD
 
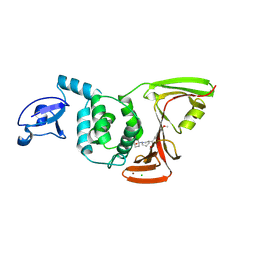 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P4 | | Descriptor: | (1S,4s)-4-{(3R)-3-[(E)-(methoxyimino)(6-methoxynaphthalen-2-yl)methyl]piperidin-1-yl}cyclohexan-1-ol, CHLORIDE ION, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYC
 
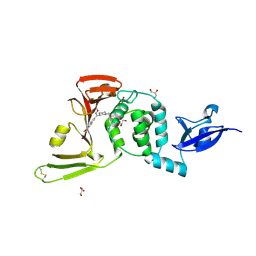 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P2 | | Descriptor: | (E)-1-[(3R)-1-cyclopentylpiperidin-3-yl]-N-methoxy-1-(6-methoxynaphthalen-2-yl)methanimine, ACETIC ACID, GLYCEROL, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchel, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
4N0B
 
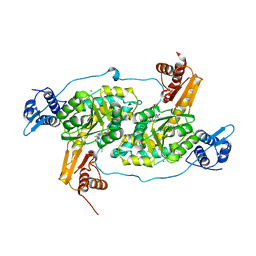 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7XEZ
 
 | |
7P2N
 
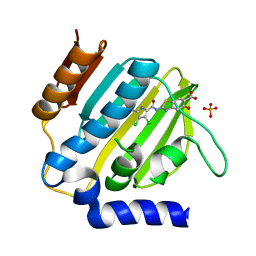 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
7M1C
 
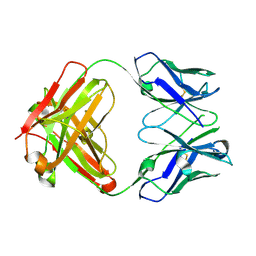 | |
7M30
 
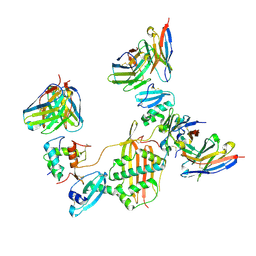 | |
7LYV
 
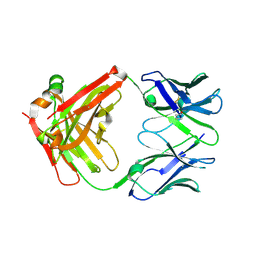 | |
7M22
 
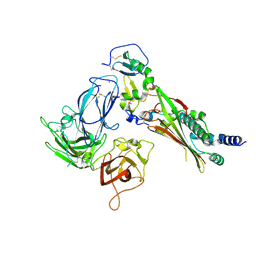 | |
7LYW
 
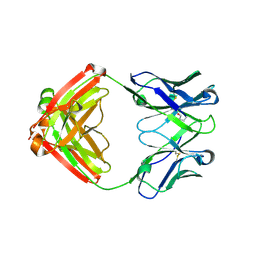 | |
2KHA
 
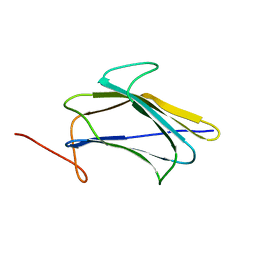 | | Solution Structure of a Pathogen Recognition Domain from a Lepidopteran Insect, Plodia interpunctella | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Dai, H, Hiromasa, Y, Fabrick, J, Vandervelde, D, Kanost, M, Krishnamoorthi, R. | | Deposit date: | 2009-03-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An initial event in the insect innate immune response: structural and biological studies of interactions between beta-1,3-glucan and the N-terminal domain of beta-1,3-glucan recognition protein
Biochemistry, 52, 2013
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
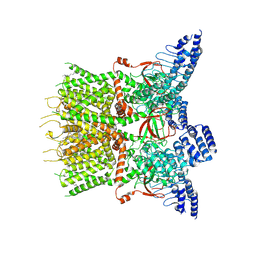 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XNK
 
 | | human KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y38
 
 | |
7XNI
 
 | | human KCNQ1-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XNN
 
 | | human KCNQ1-CaM-ML277-PIP2 complex in state B | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XNL
 
 | | human KCNQ1-CaM-ML277-PIP2 complex in state A | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6ZRN
 
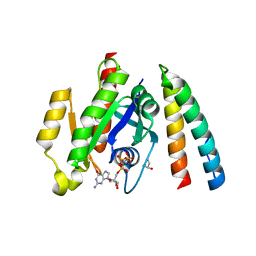 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427S/L429M/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
7ACD
 
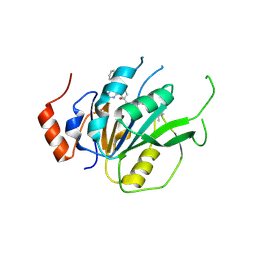 | | Crystal structure of the human METTL3-METTL14 complex with compound T30 (UZH1a) | | Descriptor: | 4-[(4,4-dimethylpiperidin-1-yl)methyl]-2-oxidanyl-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes.
Chemmedchem, 16, 2021
|
|
7A0N
 
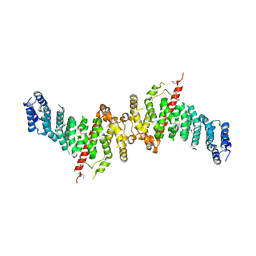 | | Structure of TSC1 NTD and linker domain | | Descriptor: | Uncharacterized protein,Uncharacterized protein | | Authors: | Fitzian, K, Kuemmel, D. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation.
Mol.Cell, 81, 2021
|
|
7A0M
 
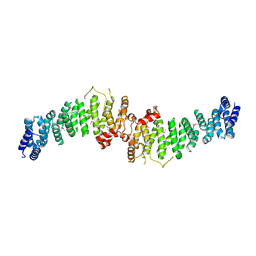 | | TSC1 N-terminal domain | | Descriptor: | SULFATE ION, TSC1 N-terminal domain | | Authors: | Zech, R, Kiontke, S, Kuemmel, D. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation.
Mol.Cell, 81, 2021
|
|
7A4F
 
 | |
