1HSQ
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | 分子名称: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | 著者 | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | 登録日 | 1994-06-13 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
3JRE
 
 | |
9UGE
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution | | 分子名称: | D-MALATE, OXALIC ACID, Peptidoglycan recognition protein 1 | | 著者 | Barik, D, Ahmad, N, Maurya, A, Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2025-04-11 | | 公開日 | 2025-05-14 | | 実験手法 | X-RAY DIFFRACTION (2.305 Å) | | 主引用文献 | Crystal structure of the complex of camel peptidoglycan recognition protein, PGRP-S with malic acid and oxalic acid at 2.3 A resolution
To Be Published
|
|
3J5R
 
 | |
1HZ2
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLEMENTARY DUPLEX 5'-D(AGGCG*CCT)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G*. MODEL OF A MALONDIALDEHYDE CROSSLINK | | 分子名称: | DNA (5'-D(*AP*GP*GP*CP*GP*CP*CP*T)-3'), PROPANE | | 著者 | Dooley, P.A, Tsarouhtsis, D, Korbel, G.A, Nechev, L.V, Shearer, J, Zegar, I.S, Harris, C.M, Stone, M.P, Harris, T.M. | | 登録日 | 2001-01-23 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural studies of an oligodeoxynucleotide containing a trimethylene interstrand cross-link in a 5'-(CpG) motif: model of a malondialdehyde cross-link.
J.Am.Chem.Soc., 123, 2001
|
|
3JV6
 
 | |
3JXD
 
 | |
3K2Y
 
 | | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B | | 分子名称: | uncharacterized protein lp_0118 | | 著者 | Seetharaman, J, Lew, S, Vorobiev, S.M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-09-30 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B
To be Published
|
|
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | 分子名称: | Dihydrofolate reductase, Nanobody | | 著者 | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | 登録日 | 2009-10-12 | | 公開日 | 2010-10-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
3KA5
 
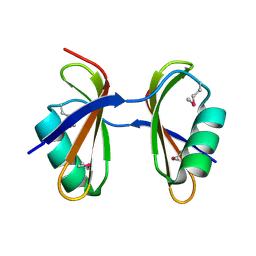 | | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum. Northeast Structural Genomics Consortium target id CaR123A | | 分子名称: | Ribosome-associated protein Y (PSrp-1) | | 著者 | Seetharaman, J, Neely, H, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-10-18 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum
To be Published
|
|
3JVC
 
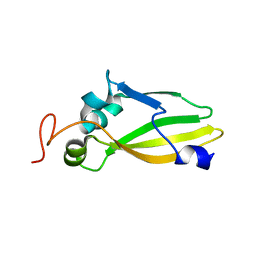 | | Crystal Structure of the Lipoprotein_17 domain from Q9PRA0_UREPA protein of Ureaplasma parvum. Northeast Structural Genomics Consortium Target UuR17a. | | 分子名称: | Conserved hypothetical membrane lipoprotein | | 著者 | Vorobiev, S, Neely, H, Lee, D, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-09-16 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal Structure of the Lipoprotein_17 domain from Q9PRA0_UREPA protein of Ureaplasma parvum.
To be Published
|
|
8AYL
 
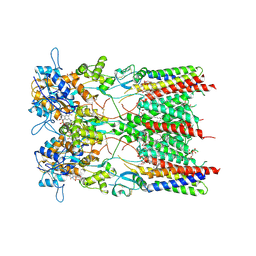 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-61432059 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 5-[2-(4-fluorophenyl)-7-(4-oxidanylpiperidin-1-yl)pyrazolo[1,5-c]pyrimidin-3-yl]-1,3-dihydroindol-2-one, ... | | 著者 | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
3JRX
 
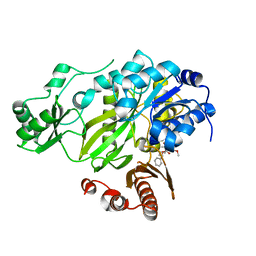 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | 分子名称: | Acetyl-CoA carboxylase 2, SORAPHEN A | | 著者 | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | 登録日 | 2009-09-09 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JYQ
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with shikimate and NADH | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Schomburg, D, Niefind, K. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
8BF8
 
 | | ISDra2 TnpB in complex with reRNA | | 分子名称: | Deinococcus radiodurans R1 chromosome 1, RNA-guided DNA endonuclease TnpB | | 著者 | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Siksnys, V, Montoya, G, Druteika, G, Silanskas, A, Venclovas, C, Karvelis, T, Kazlauskas, D. | | 登録日 | 2022-10-24 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
3JV4
 
 | |
3K1F
 
 | | Crystal structure of RNA Polymerase II in complex with TFIIB | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Kostrewa, D, Zeller, M.E, Armache, K.-J, Seizl, M, Leike, K, Thomm, M, Cramer, P. | | 登録日 | 2009-09-27 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | RNA polymerase II-TFIIB structure and mechanism of transcription initiation.
Nature, 462, 2009
|
|
3J1W
 
 | |
3J3X
 
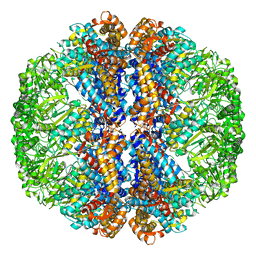 | |
3J9P
 
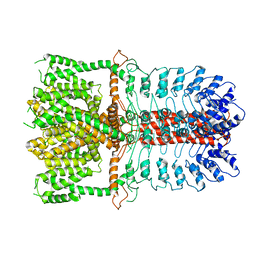 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | 分子名称: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | 著者 | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | 登録日 | 2015-02-14 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | 分子名称: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | 著者 | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | 登録日 | 2015-09-01 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
3JCG
 
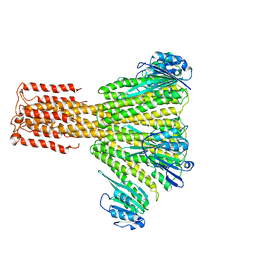 | |
3JD2
 
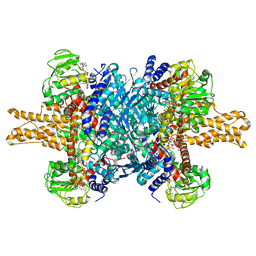 | | Glutamate dehydrogenase in complex with NADH, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | 分子名称: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | 著者 | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-09-30 | | 公開日 | 2009-10-13 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
3K73
 
 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-10-12 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
