6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | Descriptor: | Portal protein | | Authors: | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
6RRX
 
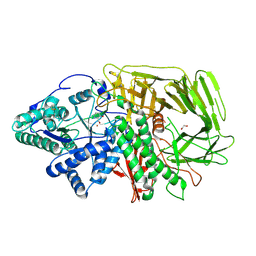 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
2N1C
 
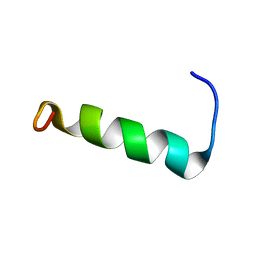 | | Structure of PvHCt, an antimicrobial peptide from shrimp litopenaeus vannamei | | Descriptor: | Hemocyanin subunit L2 | | Authors: | Petit, V.W, Rolland, J.L, Blond, A, Djediat, C, Peduzzi, J, Goulard, C, Bachere, E, Dupont, J, Destoumieux-Garzon, D, Rebuffat, S. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes.
Biochim.Biophys.Acta, 1860, 2015
|
|
2N4D
 
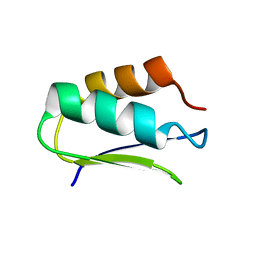 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3KFE
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
8QKB
 
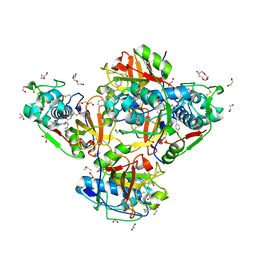 | | Crystal structure of human cathepsin L in complex with the vinyl sulfone inhibitor K777 | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-09-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
2N4B
 
 | | EC-NMR Structure of Ralstonia metallidurans Rmet_5065 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
6S59
 
 | | Structure of ovine transhydrogenase in the apo state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Nicotinamide nucleotide transhydrogenase | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2019-07-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and mechanism of mitochondrial proton-translocating transhydrogenase.
Nature, 573, 2019
|
|
6S63
 
 | | Dark-adapted structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Axford, D, Bada Juarez, J.F, Vinals, J, Watts, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two states of a light-sensitive membrane protein captured at room temperature using thin-film sample mounts.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
4D8Z
 
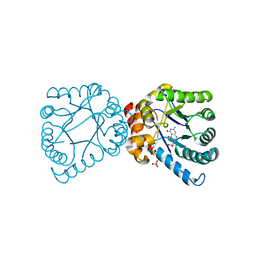 | | Crystal structure of B. anthracis DHPS with compound 24 | | Descriptor: | (3R)-3-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
3KL9
 
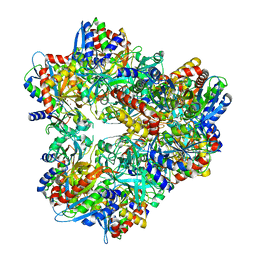 | | Crystal structure of PepA from Streptococcus pneumoniae | | Descriptor: | Glutamyl aminopeptidase, ZINC ION | | Authors: | Kim, K.K, Lee, S, Kim, D. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
2MT3
 
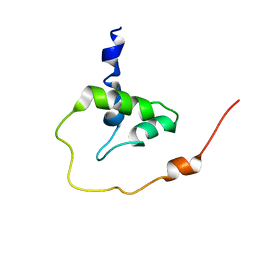 | |
6RKC
 
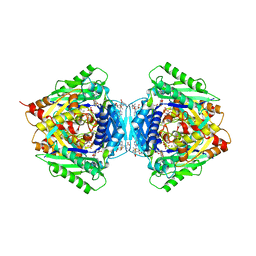 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
2N4C
 
 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
8Q4L
 
 | | GBP1 bound by 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, Guanylate-binding protein 1 | | Authors: | Pfleiderer, M.M, Liu, X, Fisch, D, Anastasakou, E, Frickel, E.M, Galej, W.P. | | Deposit date: | 2023-08-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (5.12 Å) | | Cite: | PIM1 controls GBP1 activity to limit self-damage and to guard against pathogen infection.
Science, 382, 2023
|
|
2NAJ
 
 | | Solution structure of K2 lobe of double-knot toxin | | Descriptor: | Tau-theraphotoxin-Hs1a | | Authors: | Bae, C, Anselmi, C, Kalia, J, Jara-Oseguera, A, Schwieters, C.D, Krepkiy, D, Lee, C.W, Kim, E.H, Kim, J.I, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Elife, 5, 2016
|
|
3K2T
 
 | | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A | | Descriptor: | Lmo2511 protein | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lmo2511 protein from Listeria monocytogenes, northeast structural genomics consortium target LkR84A
To be Published
|
|
3K44
 
 | | Crystal Structure of Drosophila melanogaster Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Purine-rich binding protein-alpha, ... | | Authors: | Graebsch, A, Roche, S, Niessing, D. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8QFF
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
2N6F
 
 | | Structure of Pleiotrophin | | Descriptor: | Pleiotrophin | | Authors: | Ryan, E.O, Shen, D, Wang, X. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate.
Febs J., 283, 2016
|
|
8Q6T
 
 | |
8QFG
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC after 5 seconds of blue light illumination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
7XH4
 
 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
