9BU5
 
 | | CC ProXp-ala apo solution structure | | Descriptor: | DNA-binding protein, putative | | Authors: | Duran, A.D, Foster, M.P. | | Deposit date: | 2024-05-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR-based solution structure of the Caulobacter crescentus ProXp-ala trans-editing enzyme.
Biomol.Nmr Assign., 18, 2024
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | Descriptor: | mRNA-decapping protein g5R | | Authors: | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | Deposit date: | 2020-12-10 | | Release date: | 2022-03-09 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
5T3Q
 
 | |
5UTG
 
 | | Red abalone lysin F104A | | Descriptor: | Egg-lysin | | Authors: | Wilburn, D.B, Tuttle, L.M. | | Deposit date: | 2017-02-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of sperm lysin yields novel insights into molecular dynamics of rapid protein evolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UOI
 
 | | Solution structure of the de novo mini protein HHH_rd1_0142 | | Descriptor: | HHH_rd1_0142 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
7FB8
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
5T1O
 
 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
7DMQ
 
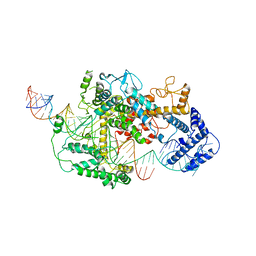 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
5TWI
 
 | |
7C96
 
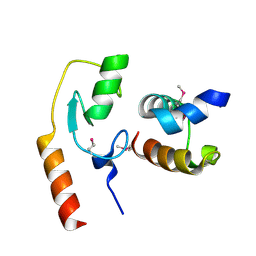 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5D14
 
 | |
5DIO
 
 | |
7FHQ
 
 | |
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
8EZV
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7OQZ
 
 | | Cryo-EM structure of human TMEM45A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Transmembrane protein 45A | | Authors: | Grieben, M, Pike, A.C.W, Evans, A, Shrestha, L, Venkaya, S, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Carpenter, E.P. | | Deposit date: | 2021-06-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | CryoEM structure of human TMEM45A
To Be Published
|
|
8F02
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a4 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
1IMT
 
 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
2ROC
 
 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
7ON1
 
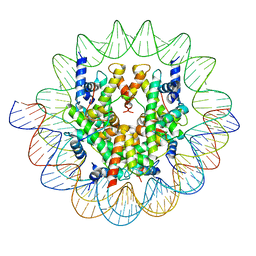 | | Cenp-A nucleosome in complex with Cenp-C | | Descriptor: | BJ4_G0006610.mRNA.1.CDS.1, BJ4_G0007000.mRNA.1.CDS.1, DNA (123-MER), ... | | Authors: | Yan, K, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cenp-A nucleosome in complex with Cenp-C
To Be Published
|
|
6JRG
 
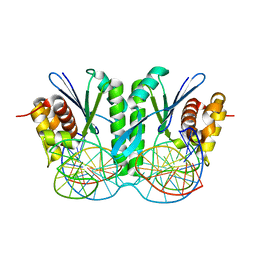 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
