4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFX
 
 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
5L8B
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
7U7H
 
 | | Cysteate acyl-ACP transferase from Alistipes finegoldii | | Descriptor: | 7-keto-8-aminopelargonate synthetase-like enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Radka, C.D, Miller, D.J, Frank, M.W, Rock, C.O. | | Deposit date: | 2022-03-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of the first step in sulfonolipid biosynthesis in Alistipes finegoldii.
J.Biol.Chem., 298, 2022
|
|
4YN0
 
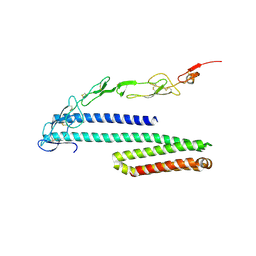 | | Crystal structure of APP E2 domain in complex with DR6 CRD domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, MAGNESIUM ION, ... | | Authors: | Xu, K, Nikolov, D. | | Deposit date: | 2015-03-08 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of DR6 in complex with the amyloid precursor protein provides insight into death receptor activation.
Genes Dev., 29, 2015
|
|
5NH4
 
 | |
5KQ4
 
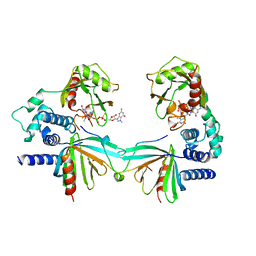 | | Crystal structure of S. pombe Dcp1/Dcp2 in complex with H. sapiens PNRC2 and synthetic cap analog | | Descriptor: | Proline-rich nuclear receptor coactivator 2, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate, mRNA decapping complex subunit 2, ... | | Authors: | Mugridge, J.S, Ziemniak, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of mRNA-cap recognition by Dcp1-Dcp2.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6RVD
 
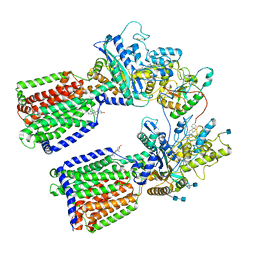 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
8UXS
 
 | |
4YJO
 
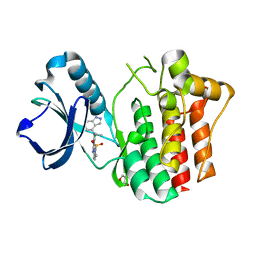 | | THE KINASE DOMAIN OF HUMAN SPLEEN TYROSINE (SYK) IN COMPLEX WITH GTC000222 | | Descriptor: | 5-chloro-N~2~-(1,1-dioxido-2,3-dihydro-1,2-benzothiazol-6-yl)-N~4~-ethyl-N~4~-(1H-indazol-4-yl)pyrimidine-2,4-diamine, GLYCEROL, Tyrosine-protein kinase SYK | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of GSK143, a highly potent, selective and orally efficacious spleen tyrosine kinase inhibitor.
Bioorg. Med. Chem. Lett., 21, 2011
|
|
5KVD
 
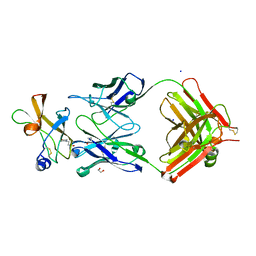 | | Zika specific antibody, ZV-2, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
7U9J
 
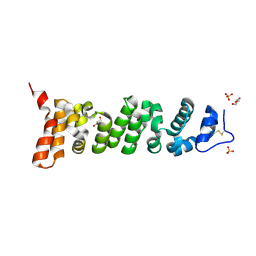 | | Crystal structure of Mesothelin-207 fragment | | Descriptor: | GLYCEROL, Isoform 3 of Mesothelin, SULFATE ION | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
6FS0
 
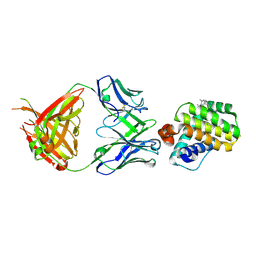 | |
7TZ6
 
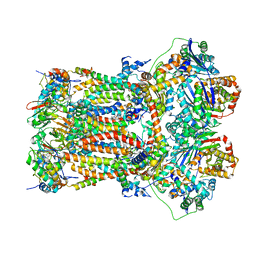 | | Structure of mitochondrial bc1 in complex with ck-2-68 | | Descriptor: | 7-chloranyl-3-methyl-2-[4-[[4-(trifluoromethyloxy)phenyl]methyl]phenyl]-1~{H}-quinolin-4-one, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Huang, R. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of complex III with bound antimalarial agent CK-2-68 provides insights into selective inhibition of Plasmodium cytochrome bc 1 complexes.
J.Biol.Chem., 299, 2023
|
|
5L2O
 
 | | Crystal Structure of ALDH1A1 in complex with BUC22 | | Descriptor: | 7-(diethylamino)-4-methyl-2H-1-benzopyran-2-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Buchman, C.D, Hurley, T.D. | | Deposit date: | 2016-08-02 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives.
J. Med. Chem., 60, 2017
|
|
7U4Z
 
 | |
6FWO
 
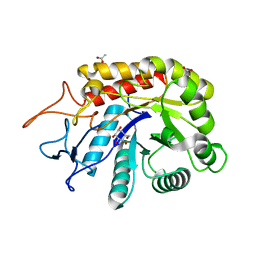 | | Structure of an E336Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
5L89
 
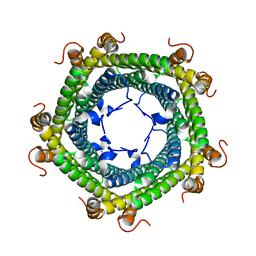 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A | | Descriptor: | CALCIUM ION, Rru_A0973 | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
4YX9
 
 | |
7LKF
 
 | | WT Chicken Scap L1-L7 / Fab 4G10 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 heavy chain, 4G10 light chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
7LKH
 
 | | Chicken Scap D435V L1-L7 domain / Fab complex focused map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4G10 Fab heavy chain, 4G10 Fab kappa chain, ... | | Authors: | Kober, D.L, Radhakrishnan, A, Goldstein, J.L, Brown, M.S, Clark, L.D, Bai, X.-C, Rosenbaum, D.M. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-30 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Scap structures highlight key role for rotation of intertwined luminal loops in cholesterol sensing.
Cell, 184, 2021
|
|
5KNZ
 
 | | Human Islet Amyloid Polypeptide Segment 19-SGNNFGAILSS-29 with Early Onset S20G Mutation Determined by MicroED | | Descriptor: | hIAPP(residues 19-29)S20G | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
7LFM
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFK
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, THR MUTANT, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFI
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE REFINED AT 1.70 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
