5L6Z
 
 | |
6QH7
 
 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
4UXX
 
 | | Structure of delta4-DgkA with AMPPCP in 9.9 MAG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Li, D, Vogeley, L, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
6E8L
 
 | | Crystal Structure of Alkyl hydroperoxidase D (AhpD) from Streptococcus pneumoniae (Strain D39/ NCTC 7466) | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Meng, Y, Davies, J, North, R, Coombes, D, Horne, C, Hampton, M, Dobson, R. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function analyses of alkylhydroperoxidase D fromStreptococcus pneumoniaereveal an unusual three-cysteine active site architecture.
J.Biol.Chem., 295, 2020
|
|
6EGK
 
 | | T181N Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
3GXE
 
 | | Complex of a Low Affinity Collagen Site with the Fibronectin 8-9FnI Domain Pair | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen alpha-1(I) chain, Fibronectin, ... | | Authors: | Sladek, B, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of collagen type I interactions with human fibronectin reveals a cooperative binding mode
J.Biol.Chem., 288, 2013
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
4ZZY
 
 | | Structure of human PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-[1-(4,4-Difluorocyclohexyl)-piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
5KNJ
 
 | | Pseudokinase Domain of MLKL bound to Compound 1. | | Descriptor: | 1-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]-3-[4-(trifluoromethyloxy)phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis.
Plos One, 11, 2016
|
|
6U3T
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6QFM
 
 | | Structure of human Mcl-1 in complex with PUMA BH3 peptide | | Descriptor: | Bcl-2-binding component 3, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6EDS
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6U66
 
 | | Structure of the trimeric globular domain of Adiponectin | | Descriptor: | Adiponectin, CALCIUM ION, SODIUM ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
8QWK
 
 | | Structure of p53 cancer mutant Y126C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Markl, A.M, Balourdas, D.I, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
6WU3
 
 | |
8GVC
 
 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
5L1W
 
 | |
3H94
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
5KNE
 
 | | CryoEM Reconstruction of Hsp104 Hexamer | | Descriptor: | Heat shock protein 104, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yokom, A.L, Gates, S.N, Jackrel, M.E, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8GVH
 
 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8QX8
 
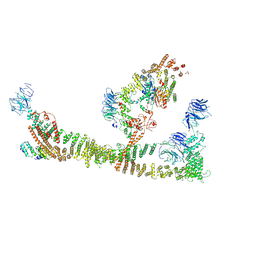 | | Endosomal membrane tethering complex CORVET | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar protein sorting-associated protein 16, ... | | Authors: | Shvarev, D, Ungermann, C, Moeller, A. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
8GY0
 
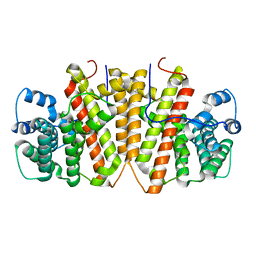 | | Agrocybe pediades linalool sunthase (Ap.LS) | | Descriptor: | Terpene synthase | | Authors: | Rehka, T, Sharma, D, Lin, F, Lim, C, Choong, Y.K, Chacko, J, Zhang, C. | | Deposit date: | 2022-09-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural Understanding of Fungal Terpene Synthases for the Formation of Linear or Cyclic Terpene Products.
Acs Catalysis, 13, 2023
|
|
6UE0
 
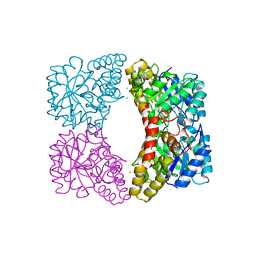 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
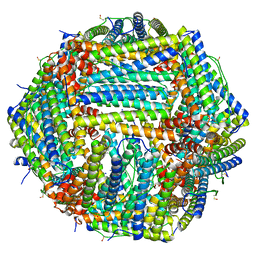
7R5O
 
 | |
