7ONF
 
 | | The binding of p-coumaroyl glucose to glycogen phosphorylase reveals the relationship between structural data and effects on cell metabolome | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Tsagkarakou, A.S, Koulas, S.M, Kyriakis, E, Drakou, C.E, Leonidas, D.D. | | Deposit date: | 2021-05-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure activity relationship of the binding of p-coumaroyl glucose to glycogen phosphorylase and its effect on hepatic cell metabolic pathways
Eur J Med Chem Rep, 3, 2021
|
|
6VRY
 
 | |
8EO5
 
 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
7PB8
 
 | | Crystal structure of the CENP-OPQUR complex | | Descriptor: | Centromere protein O, Centromere protein P, Centromere protein Q, ... | | Authors: | Bellini, D, Yatskevich, S, Muir, K.W, Barford, D. | | Deposit date: | 2021-07-31 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
8F7K
 
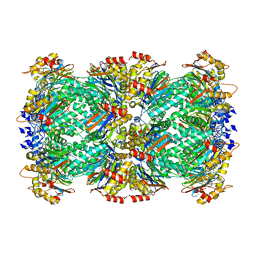 | | Thermoplasma acidophilum 20S proteasome - wild type bound to ZYA | | Descriptor: | N-[(benzyloxy)carbonyl]-L-tyrosyl-D-alanine, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Chuah, J, Smith, D. | | Deposit date: | 2022-11-18 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation.
Commun Biol, 6, 2023
|
|
5LRE
 
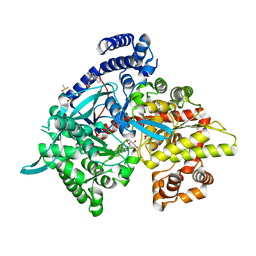 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
5LOQ
 
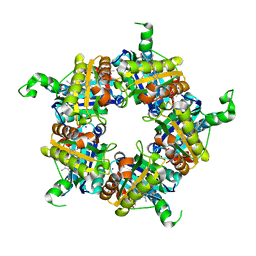 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
7P2N
 
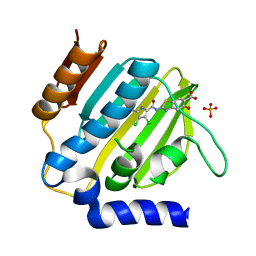 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
6WYR
 
 | | Crystal structure of anti-Muscle Specific Kinase (MuSK) Fab, MuSK1A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuSK1A heavy chain, ... | | Authors: | Vieni, C, Ekiert, D. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity maturation is required for pathogenic monovalent IgG4 autoantibody development in myasthenia gravis.
J.Exp.Med., 217, 2020
|
|
7P8P
 
 | | Crystal structure of Fhit covalently bound to a nucleotide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis(5'-adenosyl)-triphosphatase, SODIUM ION, ... | | Authors: | Herzog, D, Missun, M, Diederichs, K, Marx, A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Chemical Proteomics of the Tumor Suppressor Fhit Covalently Bound to the Cofactor Ap 3 A Elucidates Its Inhibitory Action on Translation.
J.Am.Chem.Soc., 144, 2022
|
|
8U32
 
 | | Crystal structure of PD-1 in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure- and machine learning-guided engineering demonstrate that a non-canonical disulfide in an anti-PD-1 rabbit antibody does not impede antibody developability.
Mabs, 16, 2024
|
|
6NSM
 
 | |
8GD4
 
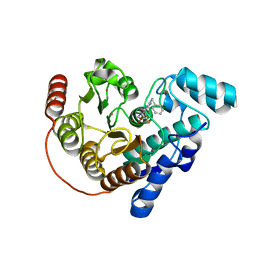 | | Crystal Structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with DMFO Inhibitor 6 | | Descriptor: | 2-(benzylamino)-N'-(difluoroacetyl)pyrimidine-5-carbohydrazide, Hdac6 protein, POTASSIUM ION, ... | | Authors: | Watson, P.R, Craigin, A.D, Christianson, D.W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Difluoromethyl-1,3,4-oxadiazoles Are Selective, Mechanism-Based, and Essentially Irreversible Inhibitors of Histone Deacetylase 6 .
J.Med.Chem., 66, 2023
|
|
6NSR
 
 | |
8G63
 
 | |
8G97
 
 | |
8UTF
 
 | |
5NFV
 
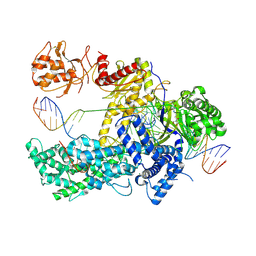 | | Crystal structure of catalytically inactive FnCas12 mutant bound to an R-loop structure containing a pre-crRNA mimic and full-length DNA target | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CRISPR-associated endonuclease Cpf1, DNA non-target strand, ... | | Authors: | Swarts, D.C, van der Oost, J, Jinek, M. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Mol. Cell, 66, 2017
|
|
5NH9
 
 | |
8G66
 
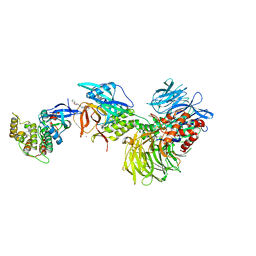 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
8G1T
 
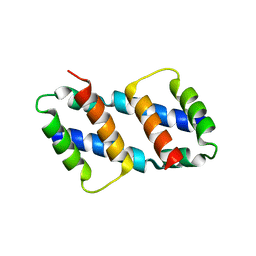 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P21 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-02-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8G4Z
 
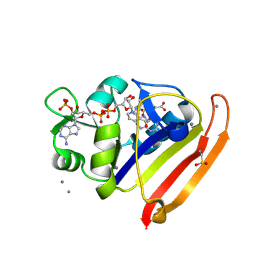 | | E. coli DHFR complex with NADP+ and folate: EF-X off model by Laue diffraction (no electric field) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8G50
 
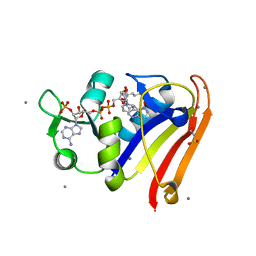 | | E. coli DHFR complex with NADP+ and folate: EF-X excited state model by Laue diffraction (electric field along b axis; 8-fold extrapolation of structure factor differences) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Klureza, M.A, Hekstra, D.R. | | Deposit date: | 2023-02-10 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5MW6
 
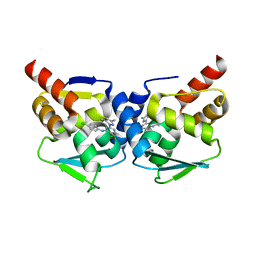 | | Crystal structure of the BCL6 BTB-domain with compound 1 | | Descriptor: | 5-chloranyl-~{N}-(4-chlorophenyl)-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-amine, B-cell lymphoma 6 protein | | Authors: | Davies, D.R, Kessler, D. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
7RM1
 
 | |
